Section 6 SNP decomposition
# devtools::install_github('philippmuench/HaplotypeDeconstructor')
data(omm_snp_annotation)
source("nmf_functions.R")
data(omm_metadata)
omm_metadata$group2 <- paste0("d", omm_metadata$day, "-", omm_metadata$mouse)
dat <- readRDS("data/rds/omm_ab.rds")
datre <- readRDS("data/rds/reseq.rds")
create_df <- function(dat = dat, datre = datre, bug = bug) {
dat <- dat[which(dat$chr == bug), ]
dat$snp_id <- paste0(dat$alteration, " ", dat$POS)
dat$sample <- paste0("AB study ", dat$mouse.id, " d", dat$day, " ", group = dat$group, sample = dat$mouse.group)
df <- data.frame(id = dat$snp_id, AF = dat$AF, sample = dat$sample)
datre <- datre[which(datre$chr == bug), ]
if (nrow(datre) > 0) {
message(nrow(datre))
datre$snp_id <- paste0(datre$alteration, " ", datre$POS)
datre$sample <- paste0("reseq ", datre$mouse.id)
dfre <- data.frame(id = datre$snp_id, AF = datre$AF, sample = datre$sample)
df <- rbind(df, dfre)
}
# prepare data
omm <- tidyr::spread(df, sample, AF)
omm[is.na(omm)] <- 0
rownames(omm) <- omm$id
omm$id <- NULL
return(omm)
}6.1 Blautia_coccoides
## 226# gof <- assessNumberHaplotyes(data.matrix(omm), 2:8, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 3)
plotHaplotypeMap(decomposed)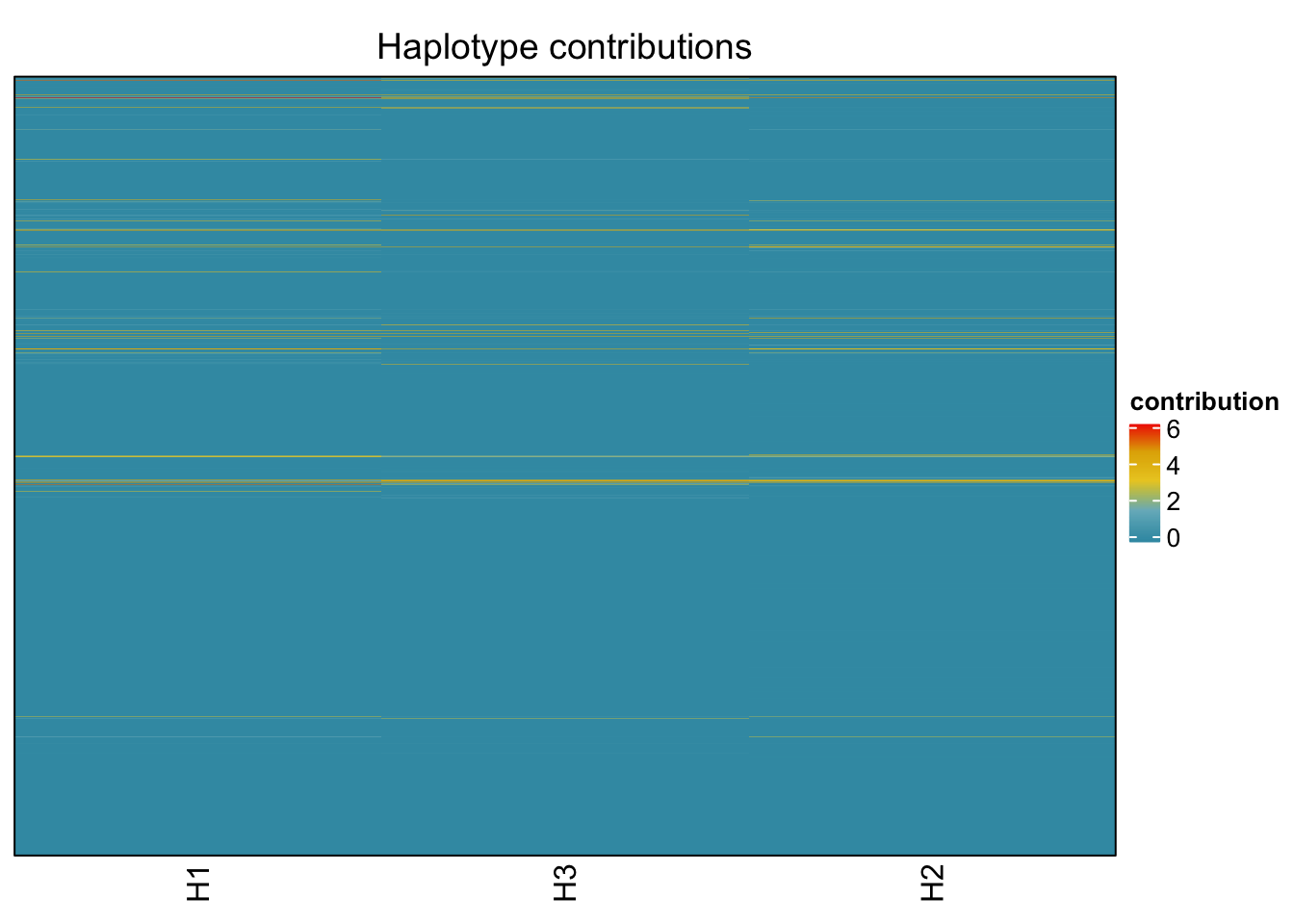
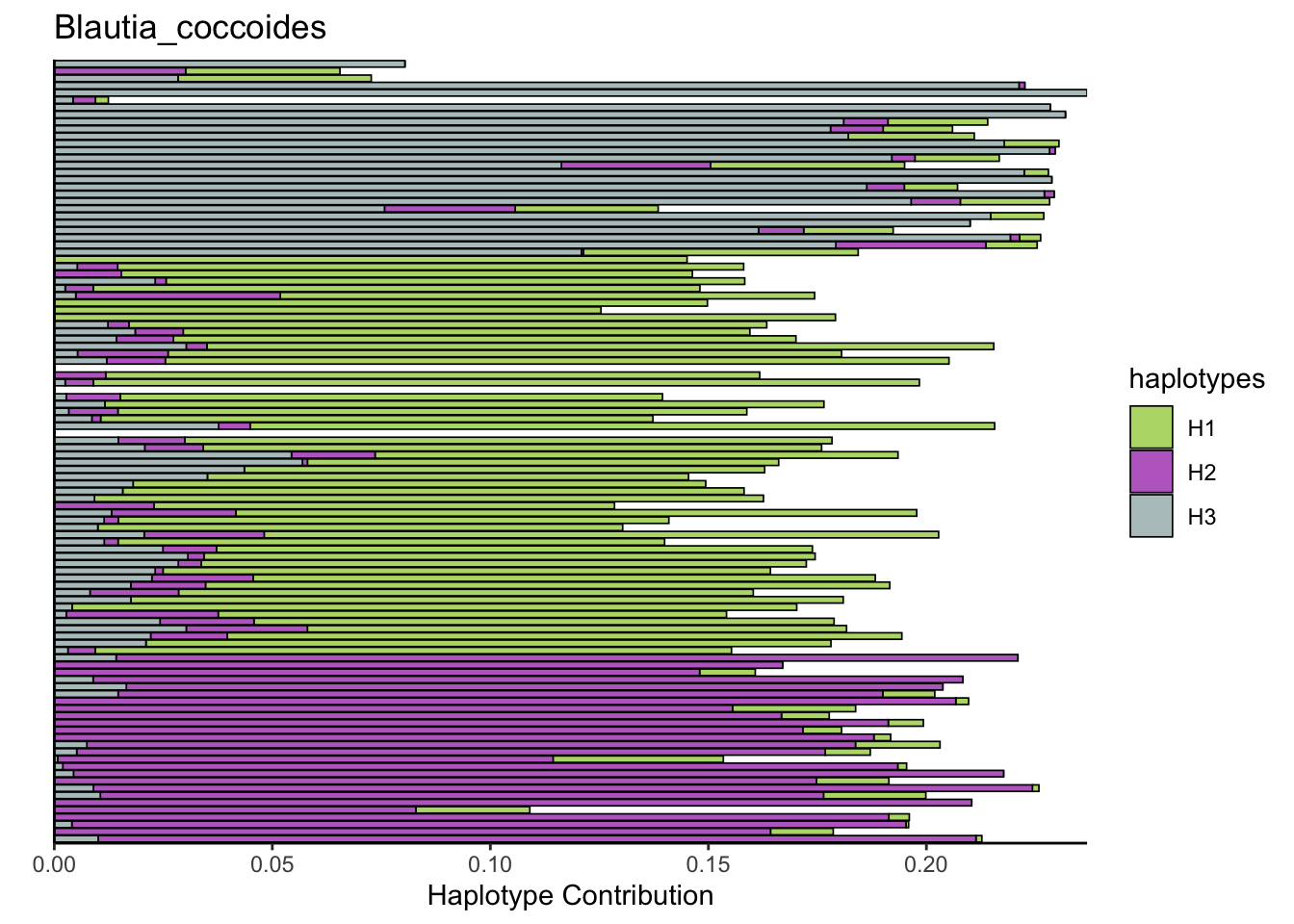
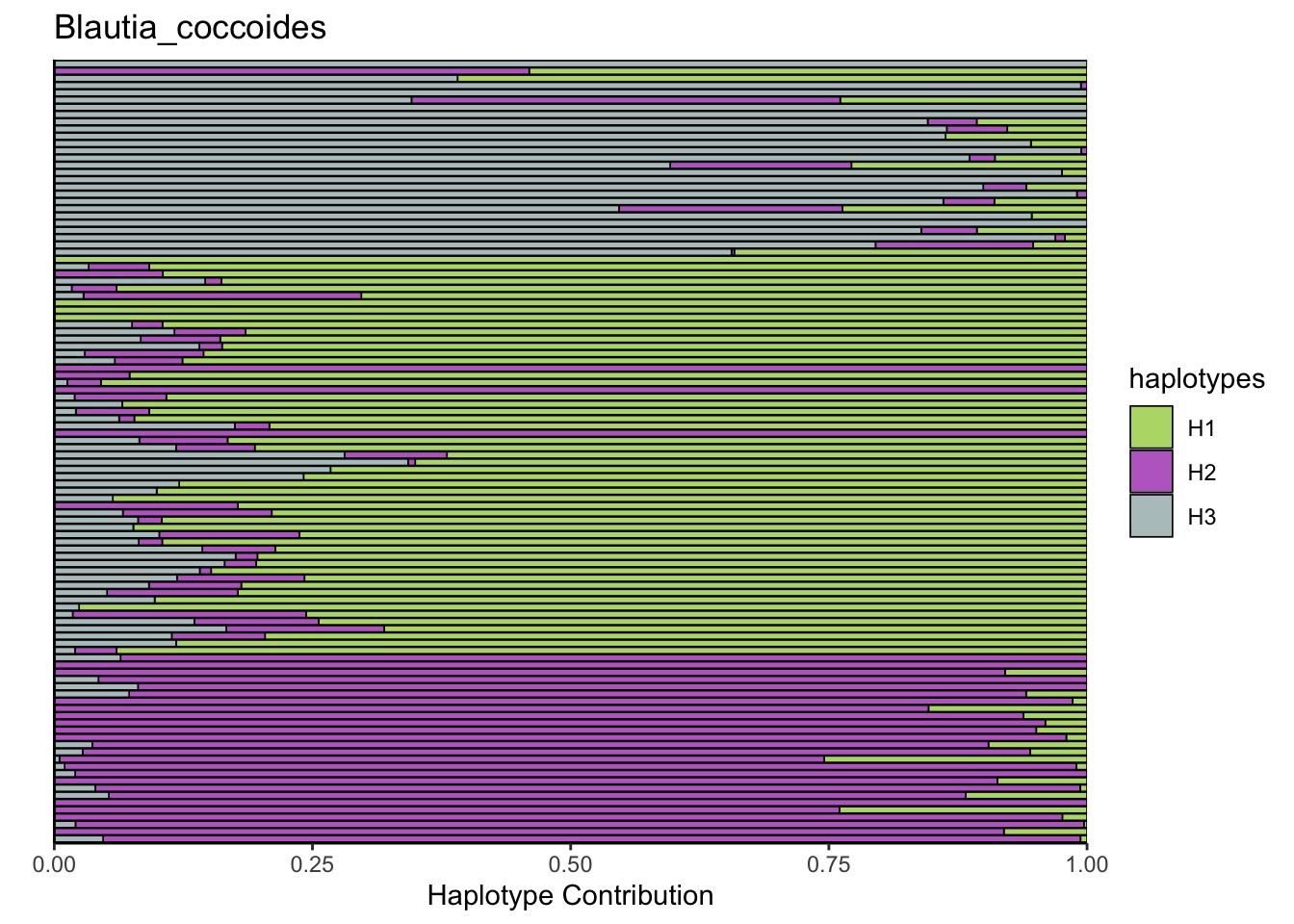
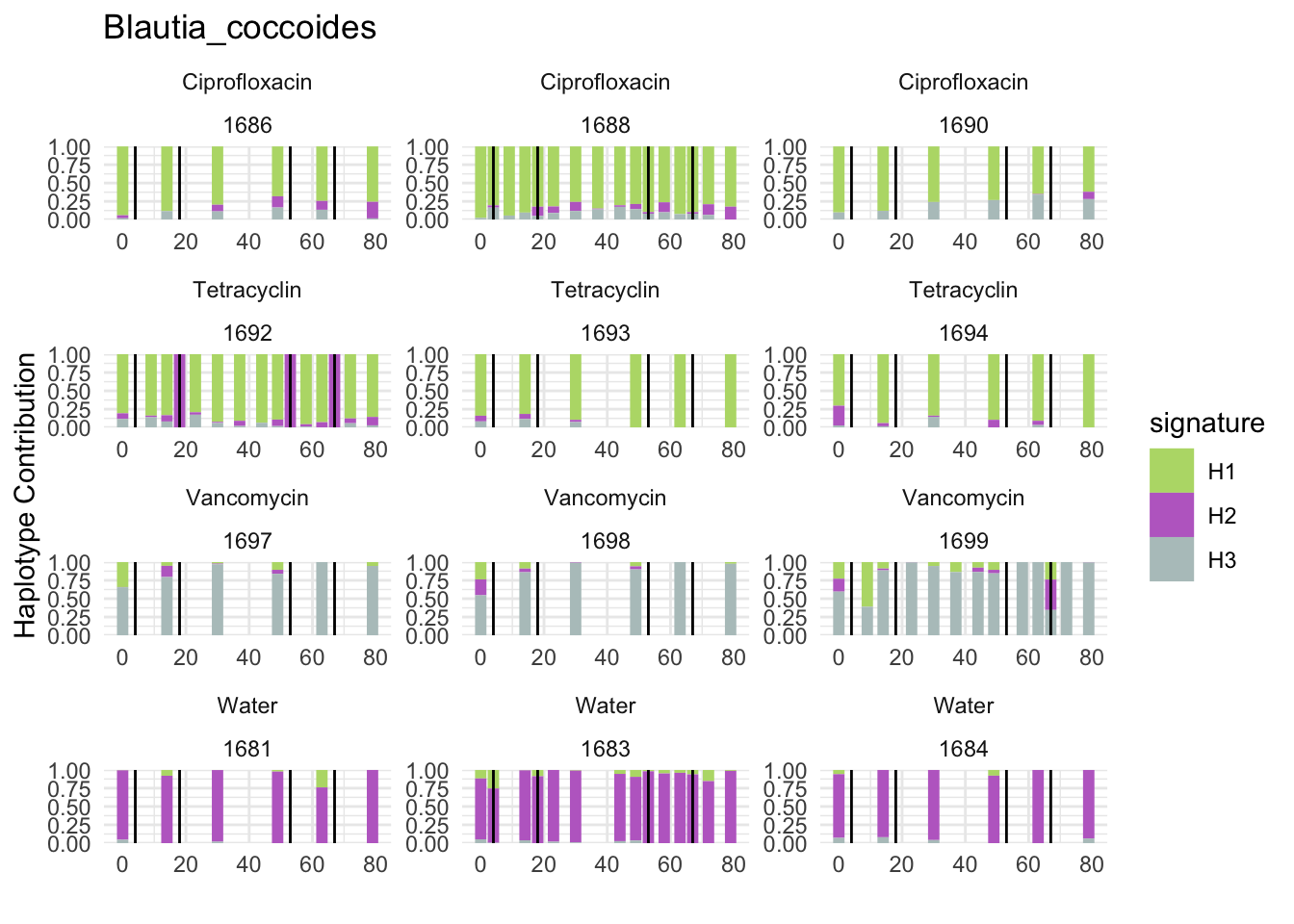
6.2 Akkermansia_muciniphila
bug <- "Akkermansia_muciniphila"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 6)
plotHaplotypeMap(decomposed)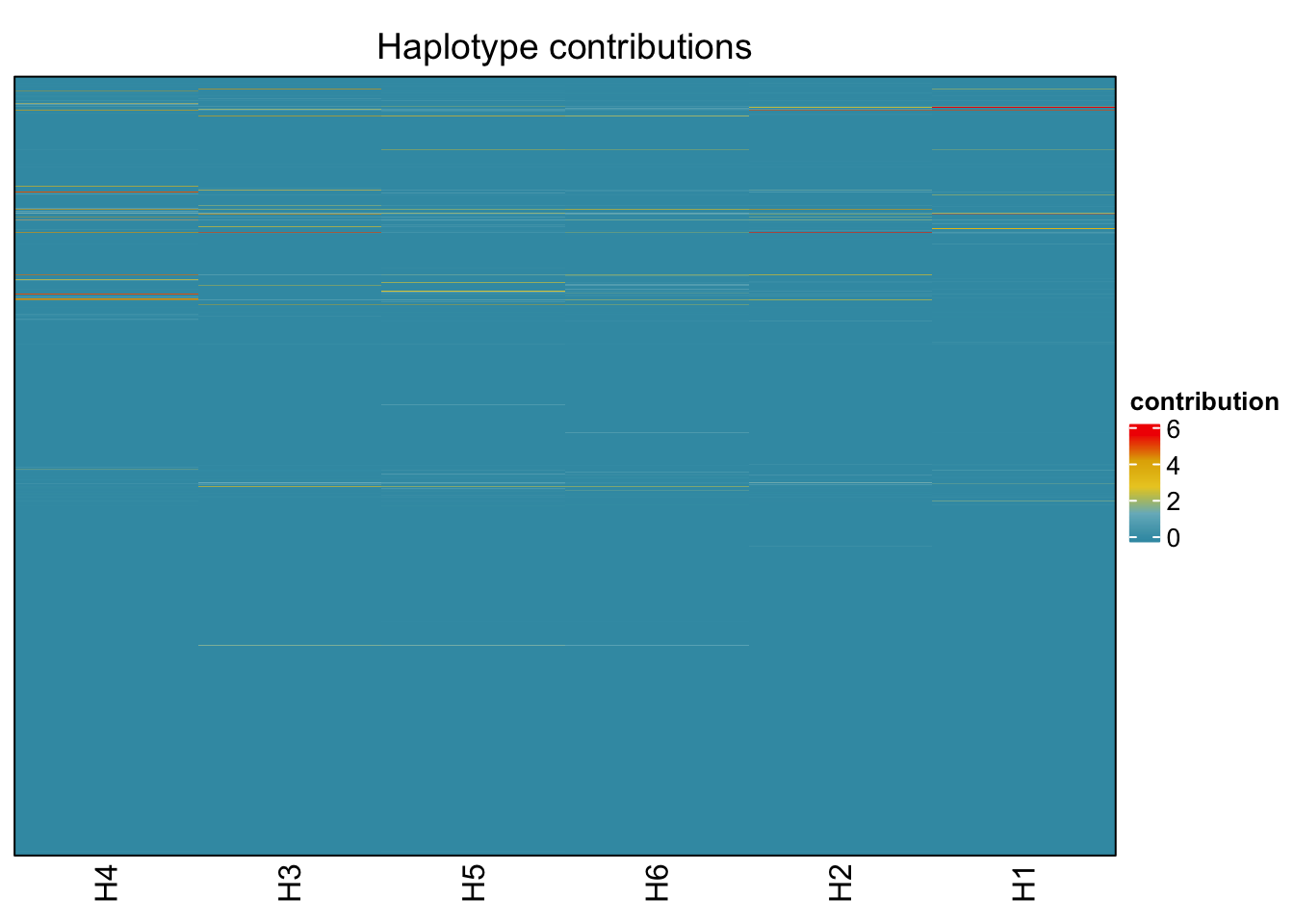
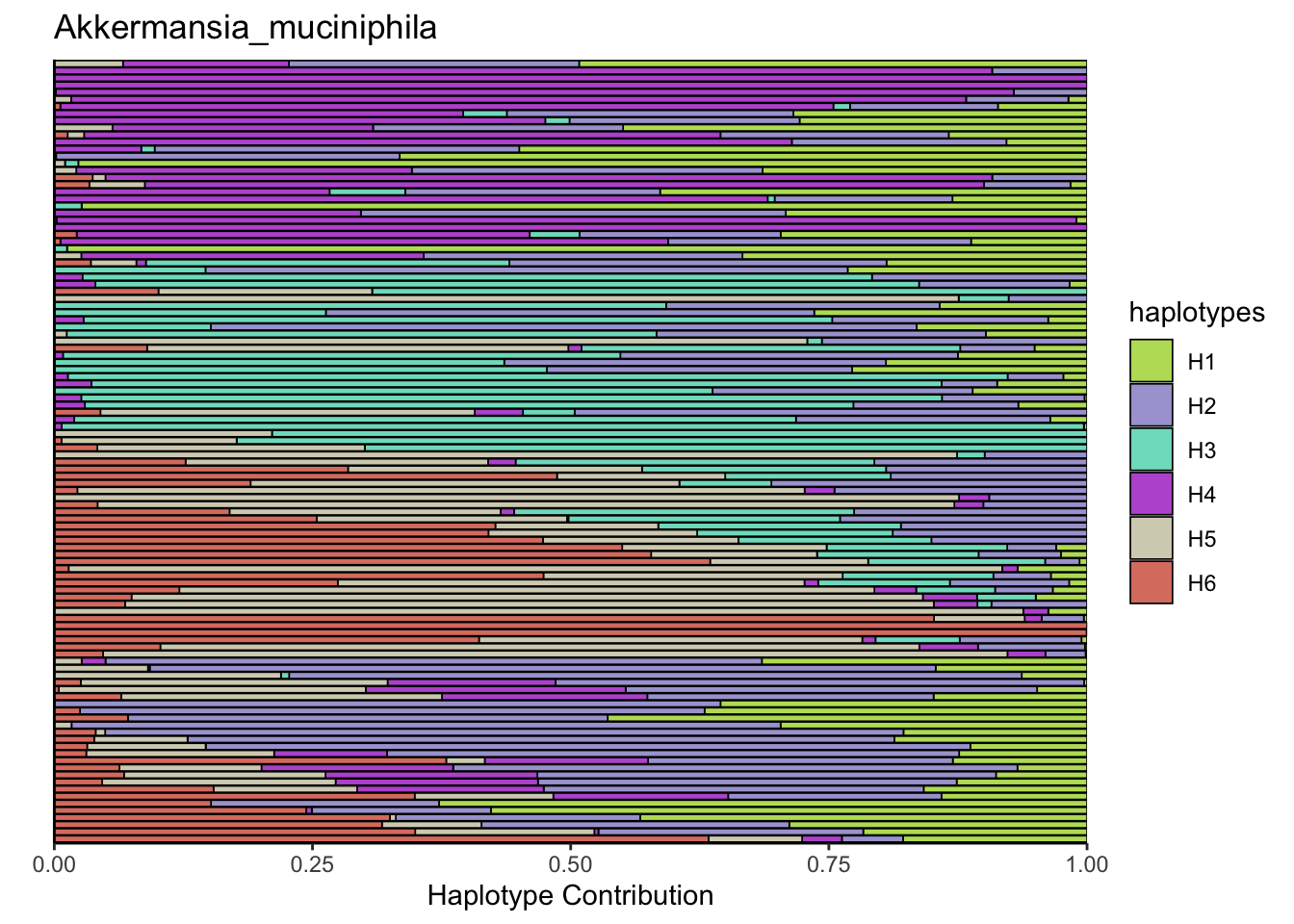
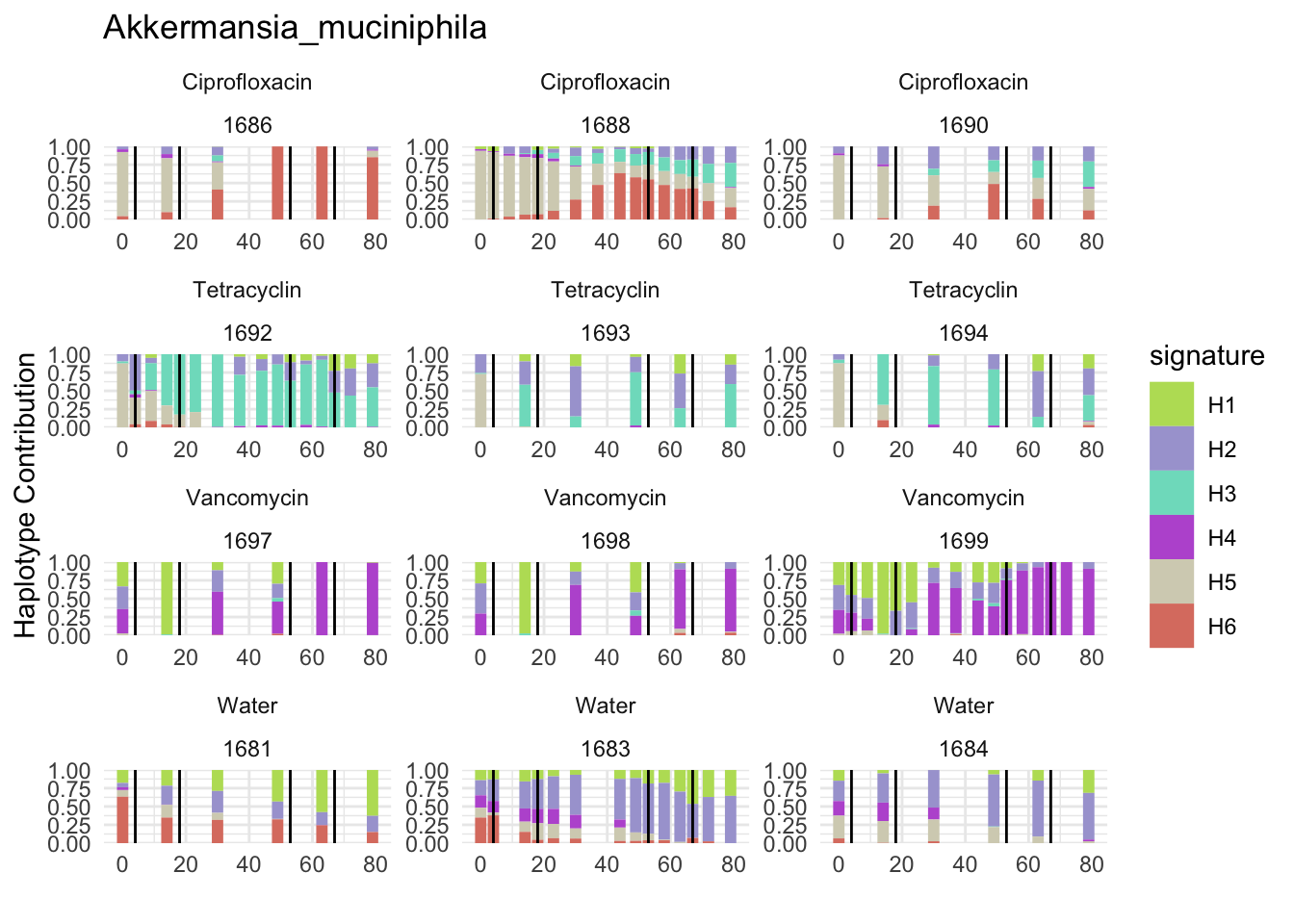
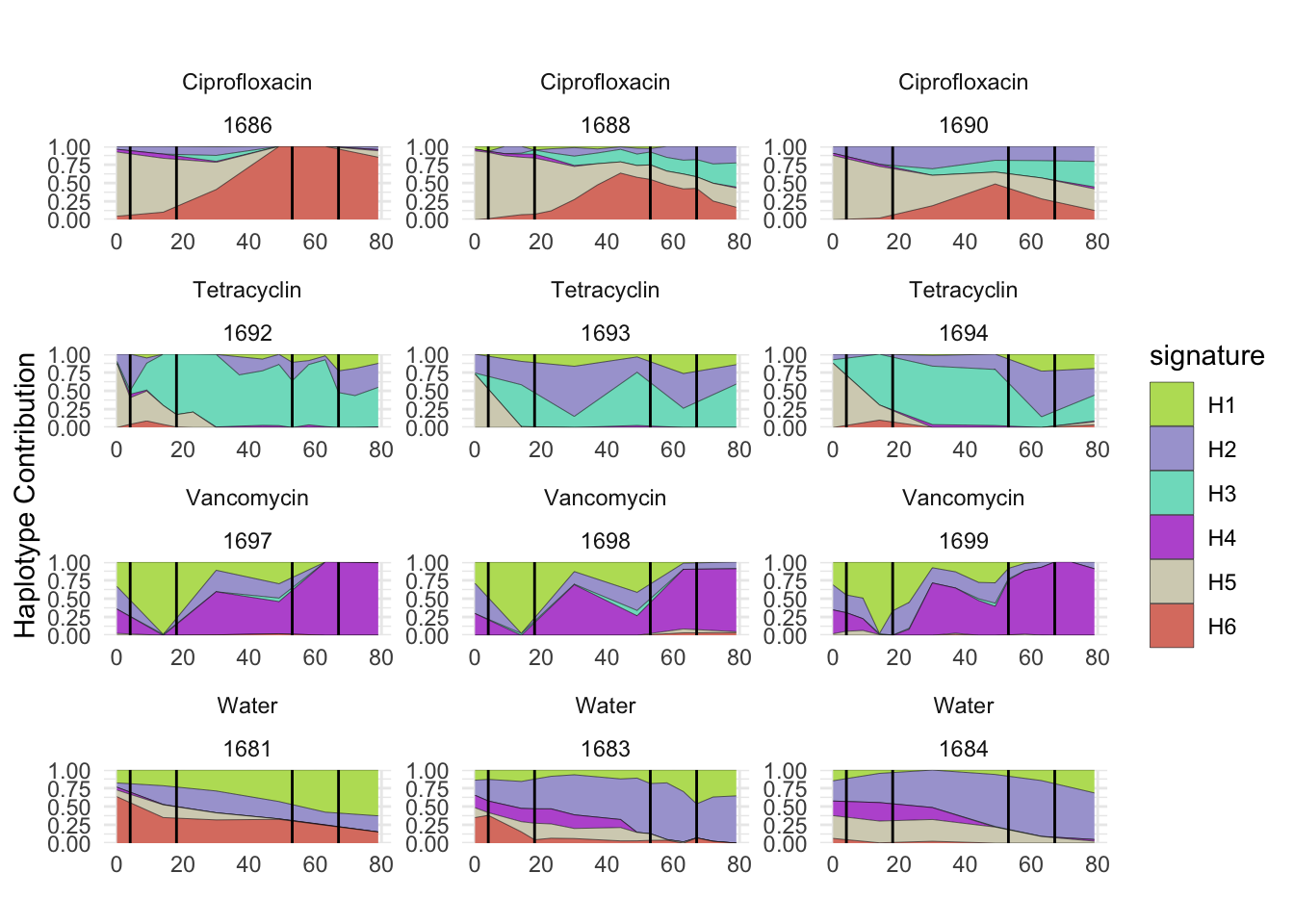
6.3 Clostridioforme
bug <- "Clostridioforme"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 4)
plotHaplotypeMap(decomposed)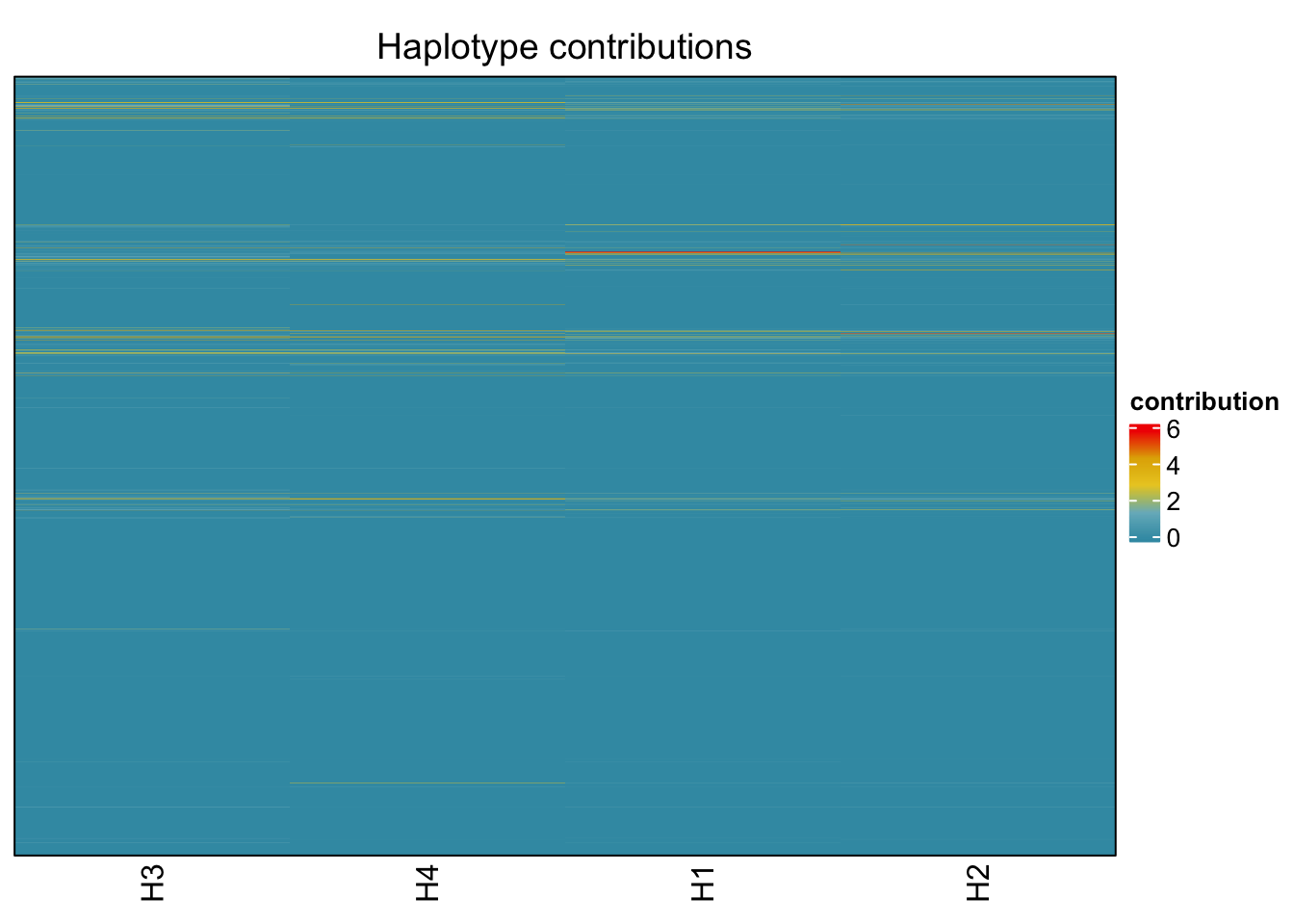
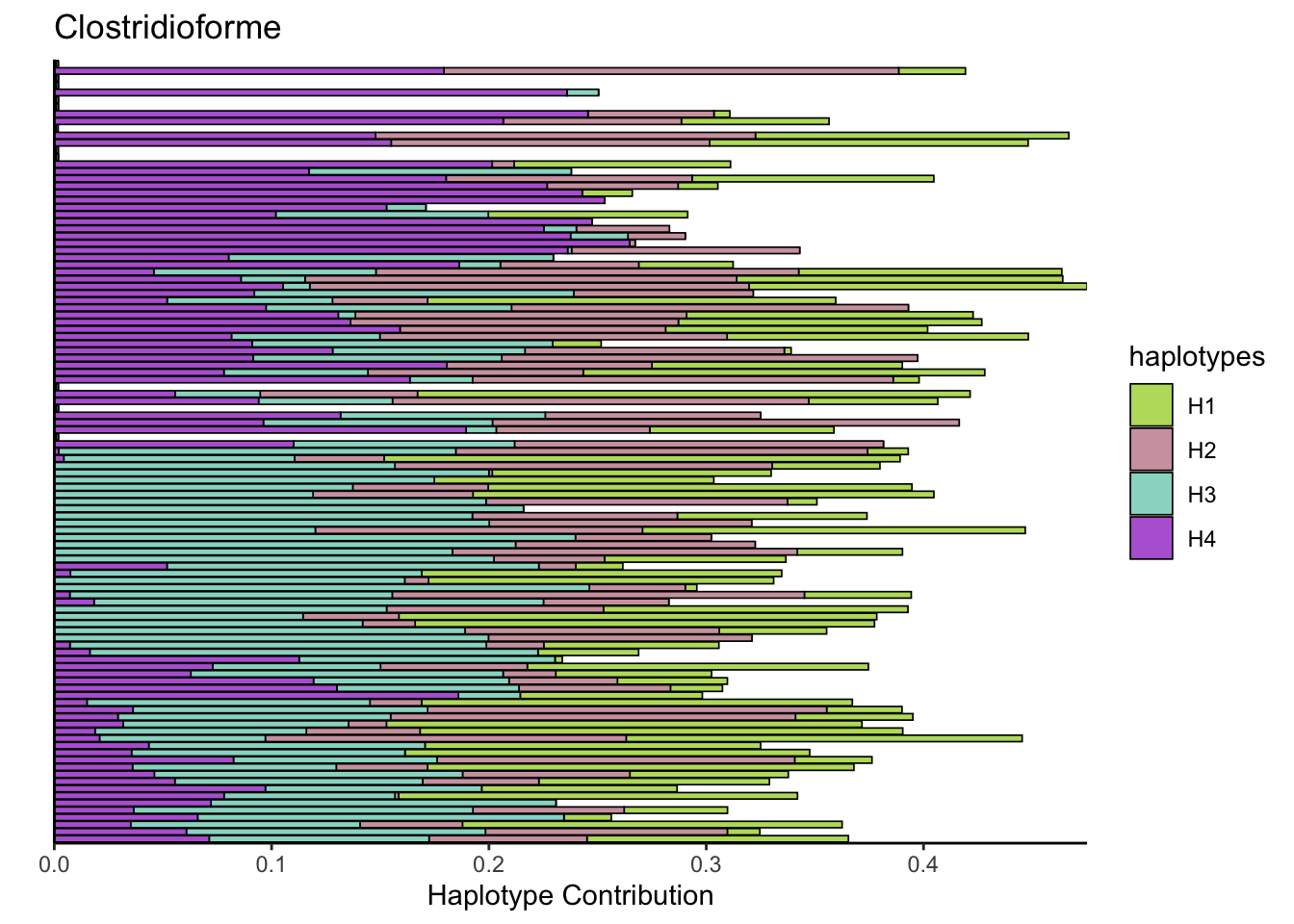
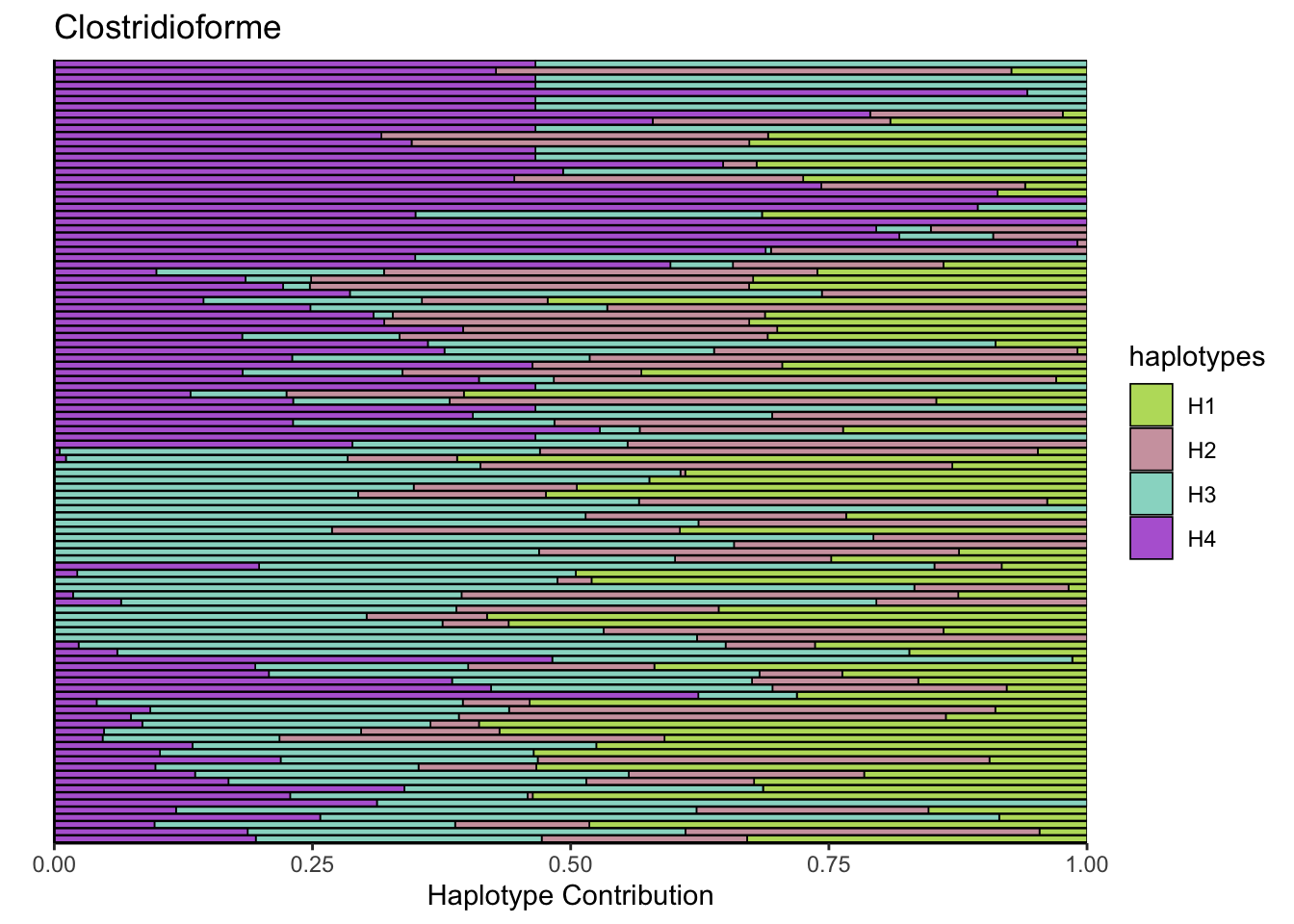
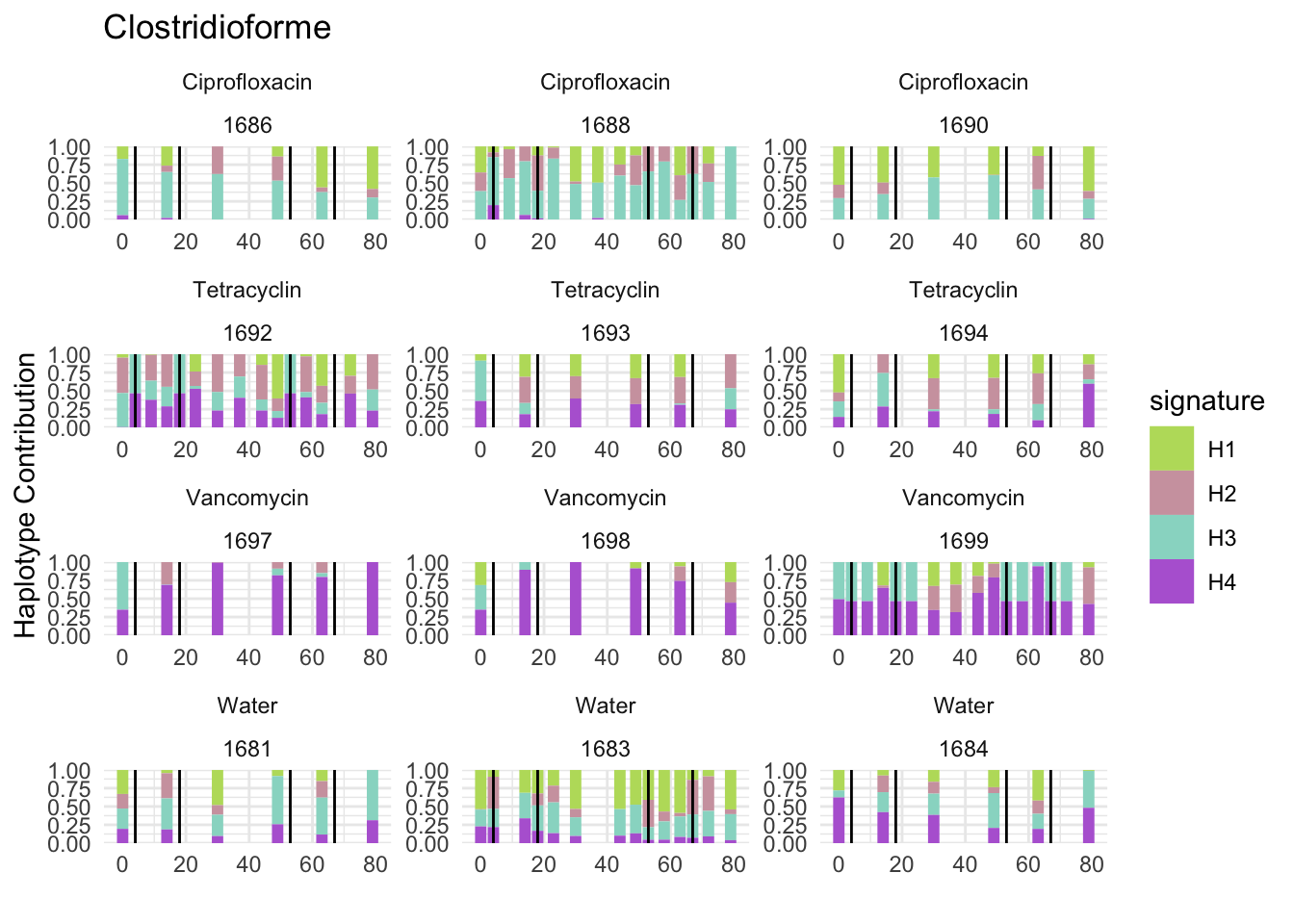
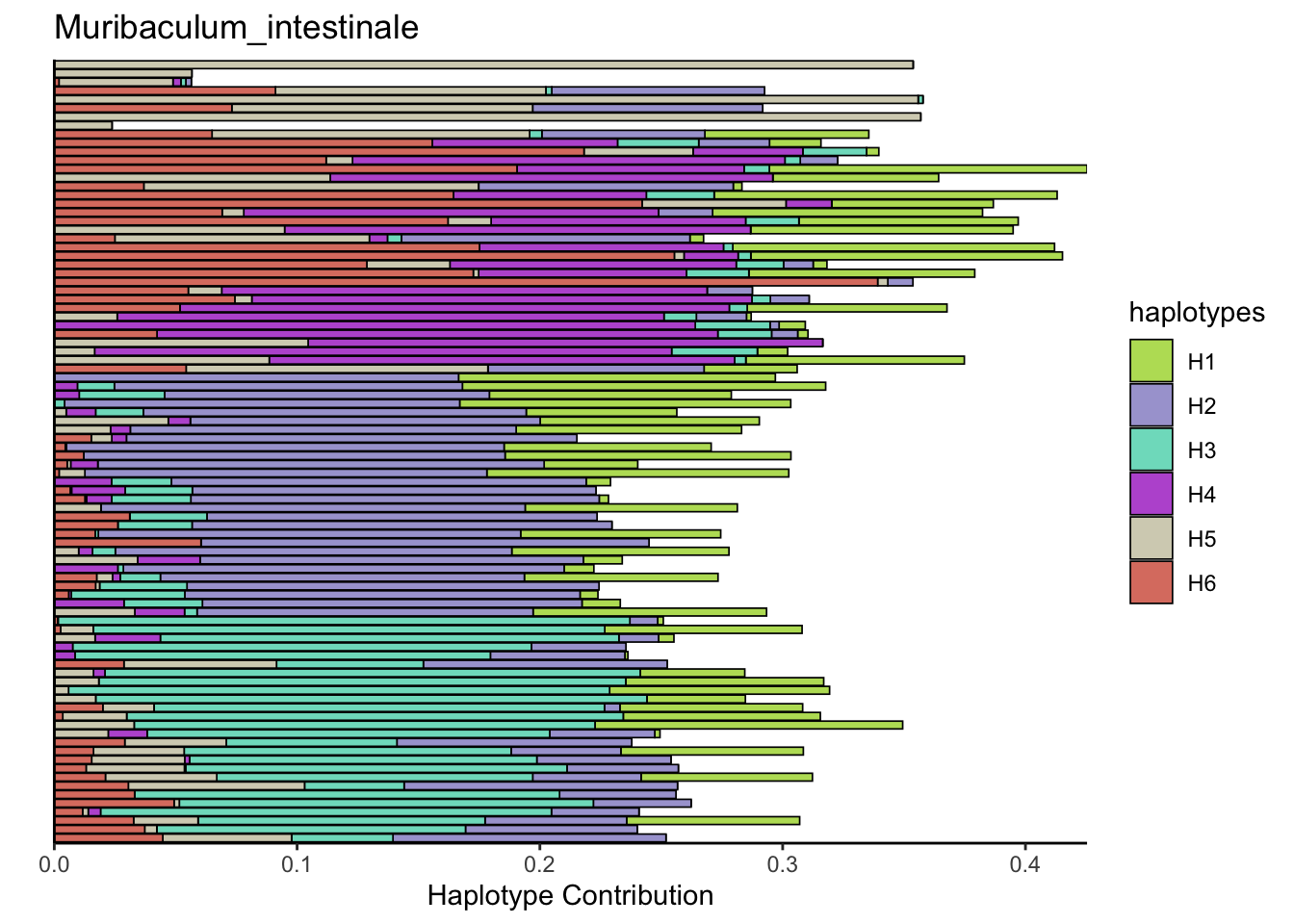
 ## Muribaculum_intestinale
## Muribaculum_intestinale
bug <- "Muribaculum_intestinale"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 6)
plotHaplotypeMap(decomposed)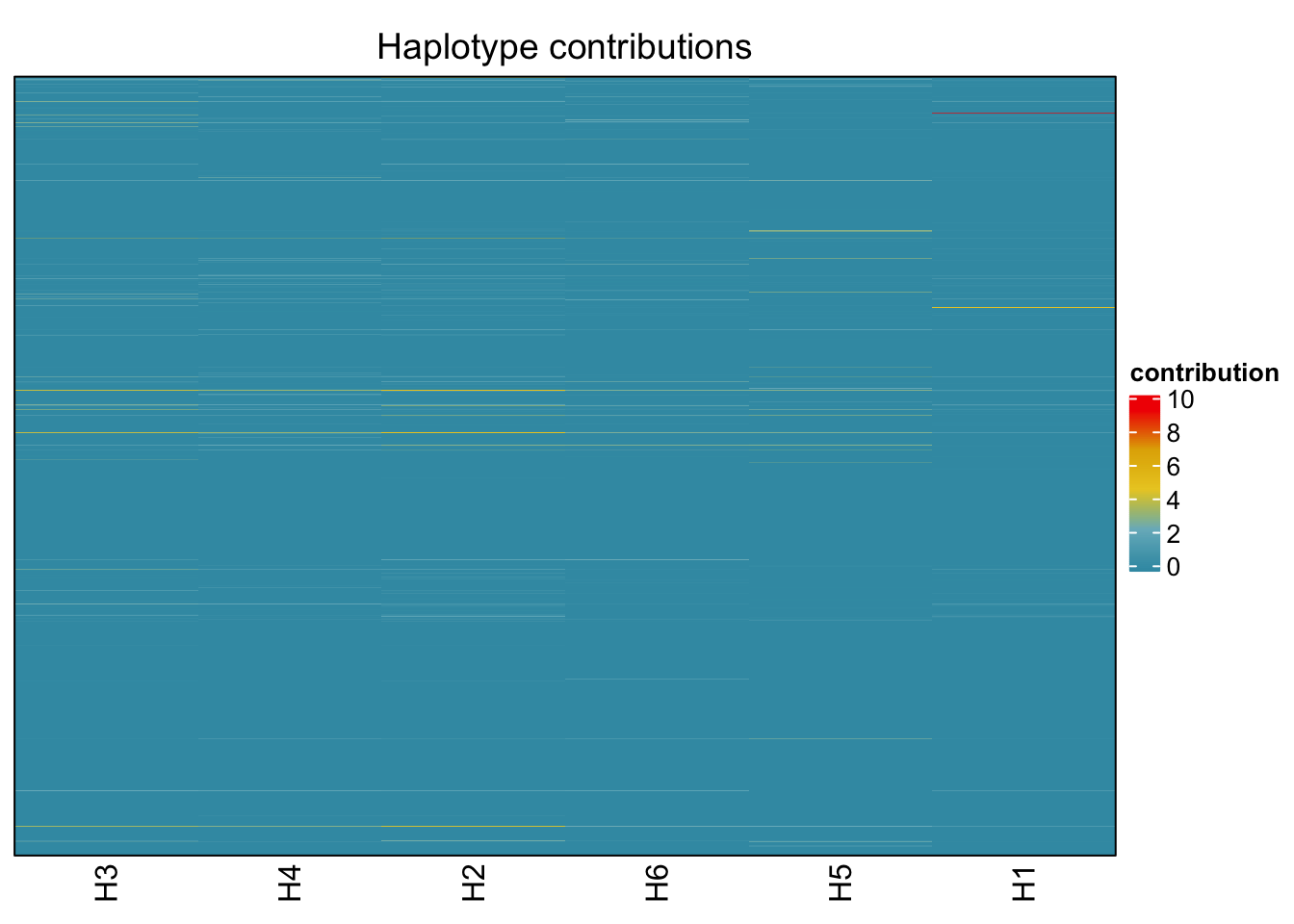
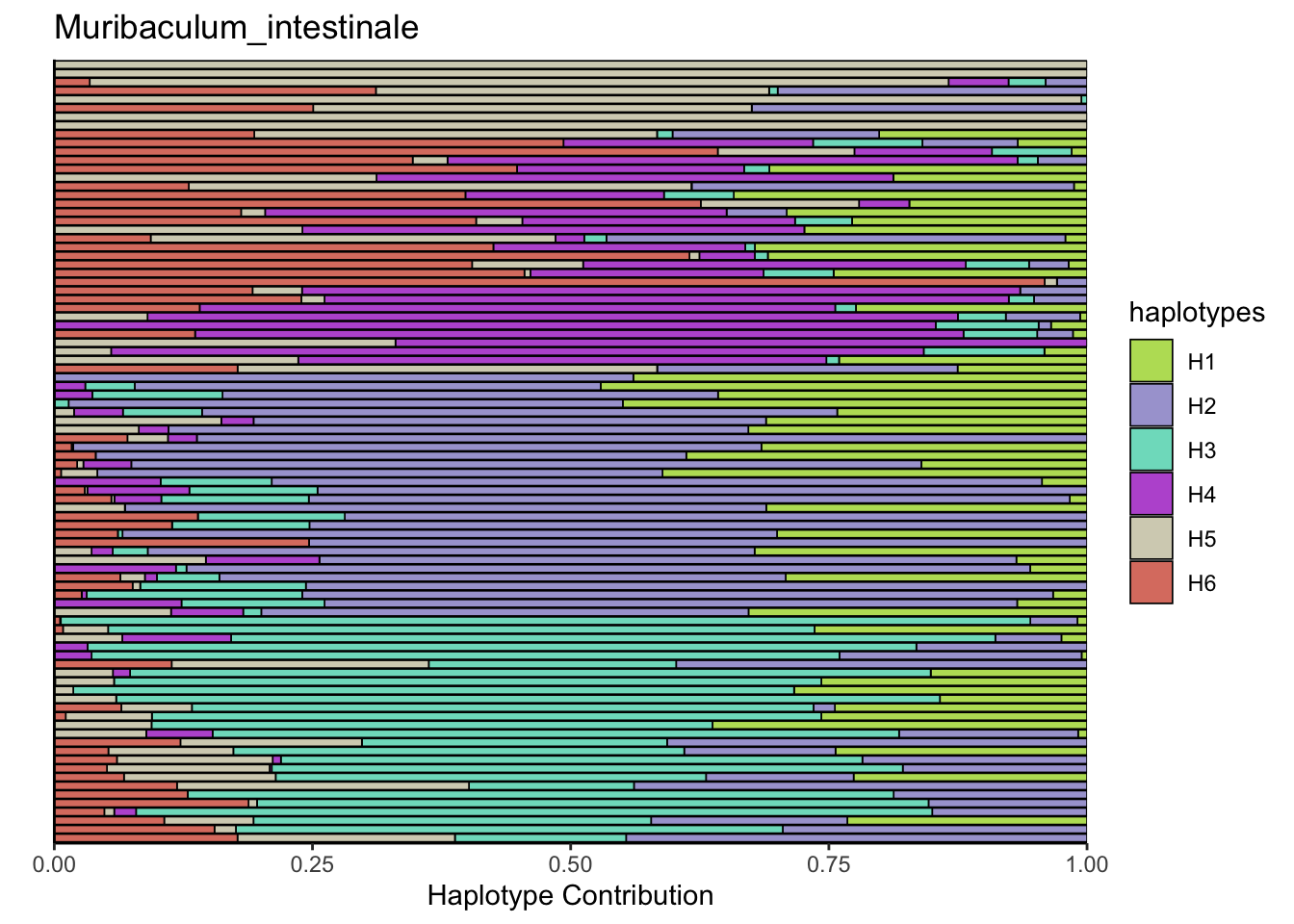
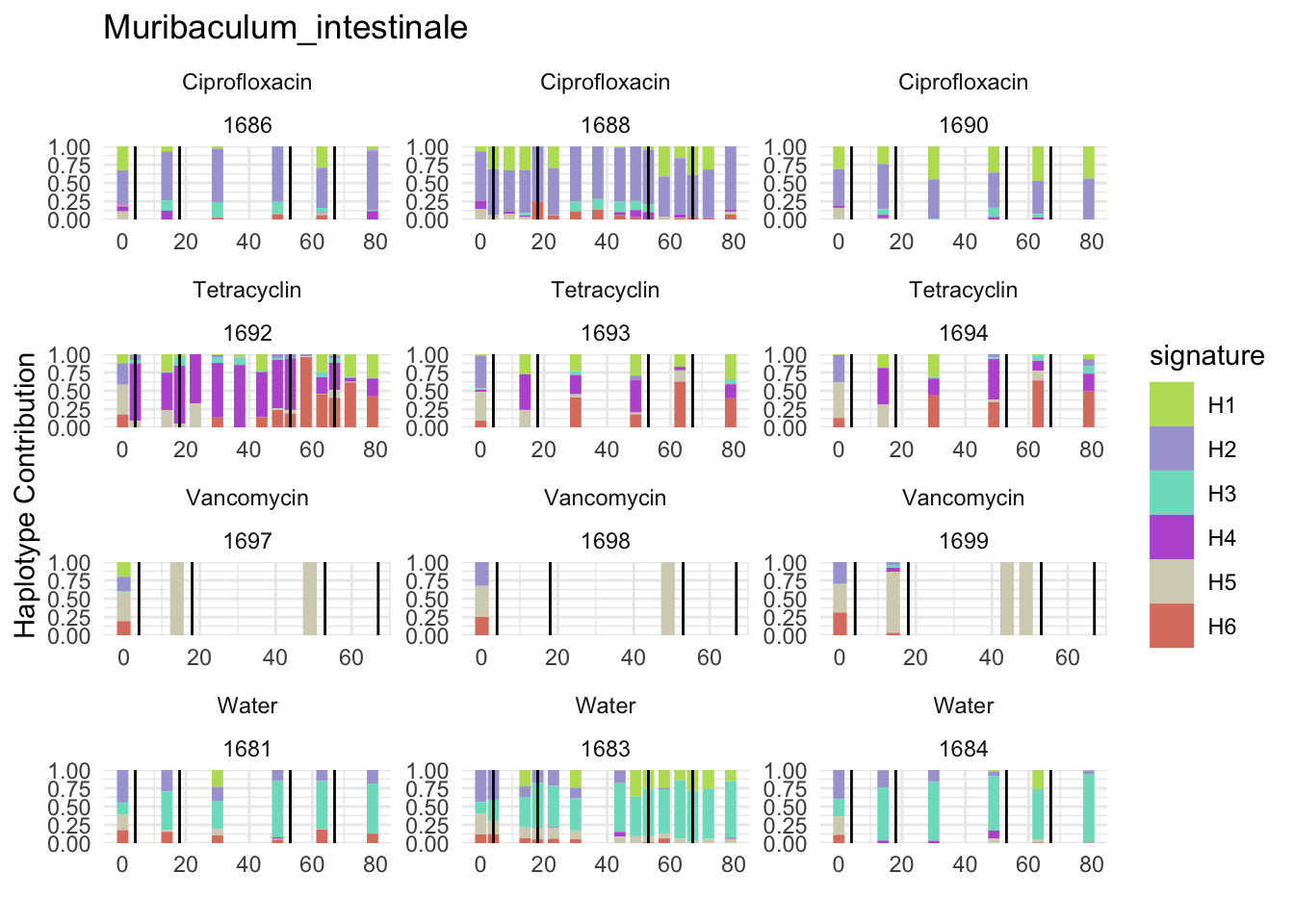
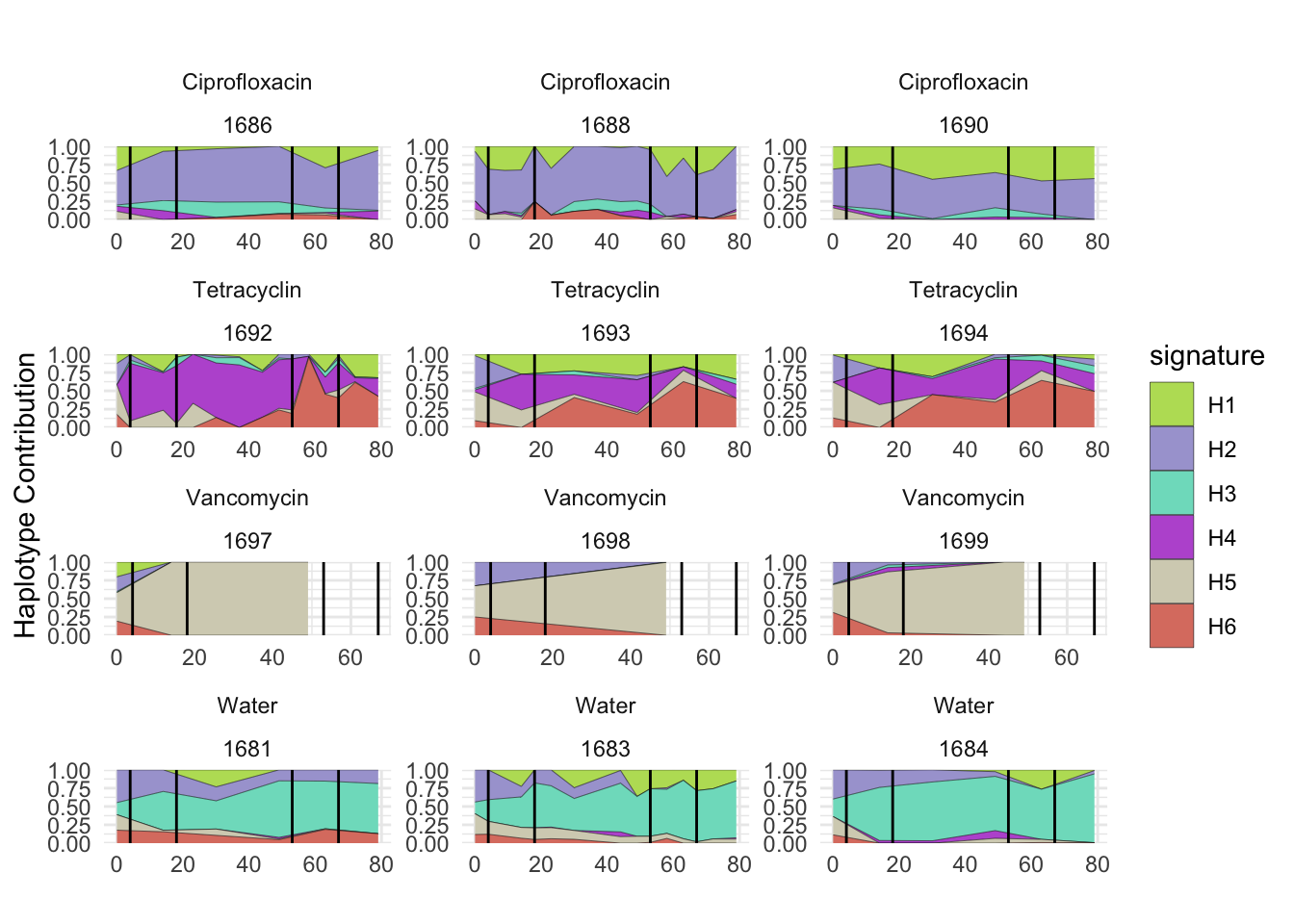
 ## T_muris
## T_muris
bug <- "T_muris"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 4)
plotHaplotypeMap(decomposed)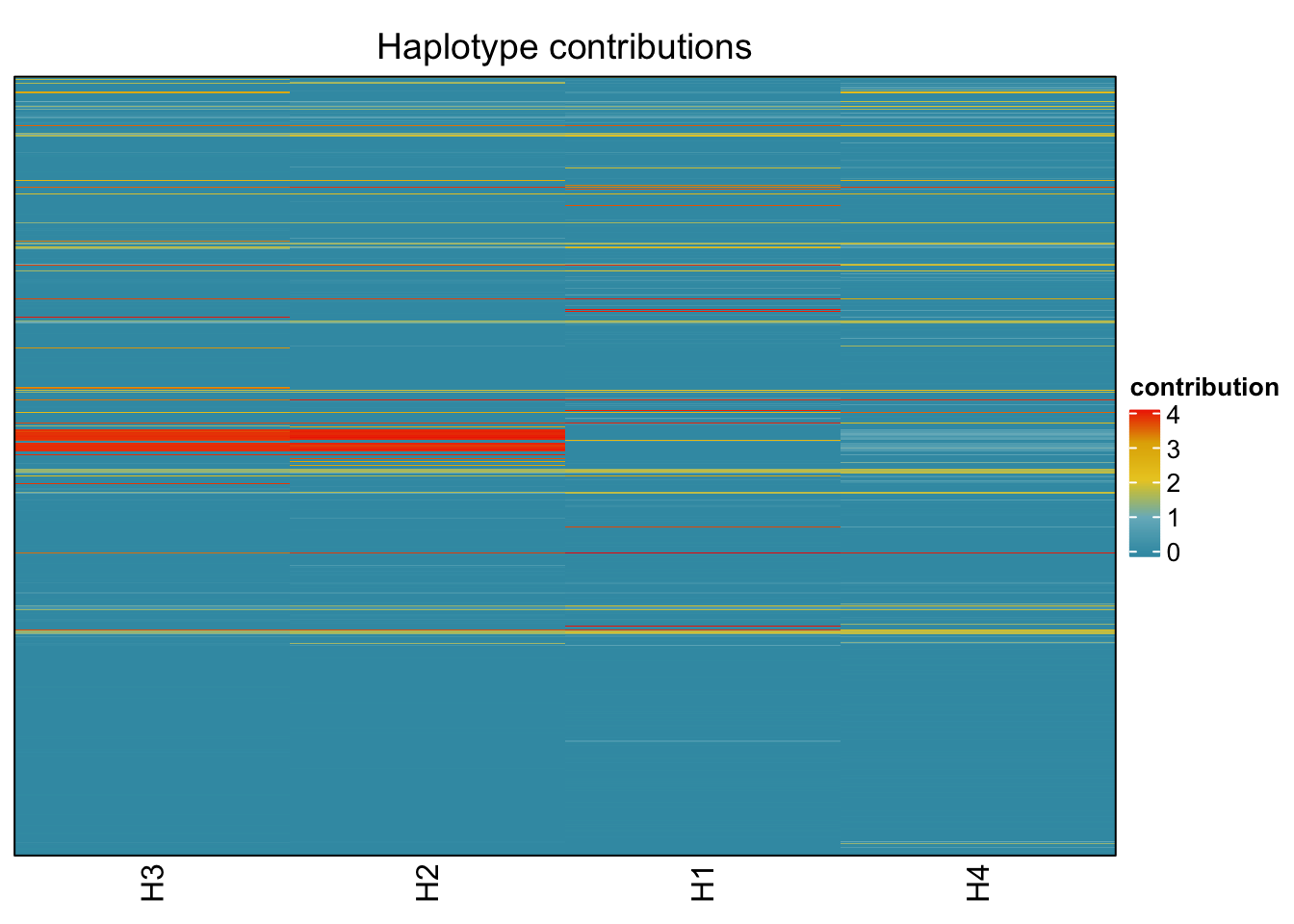
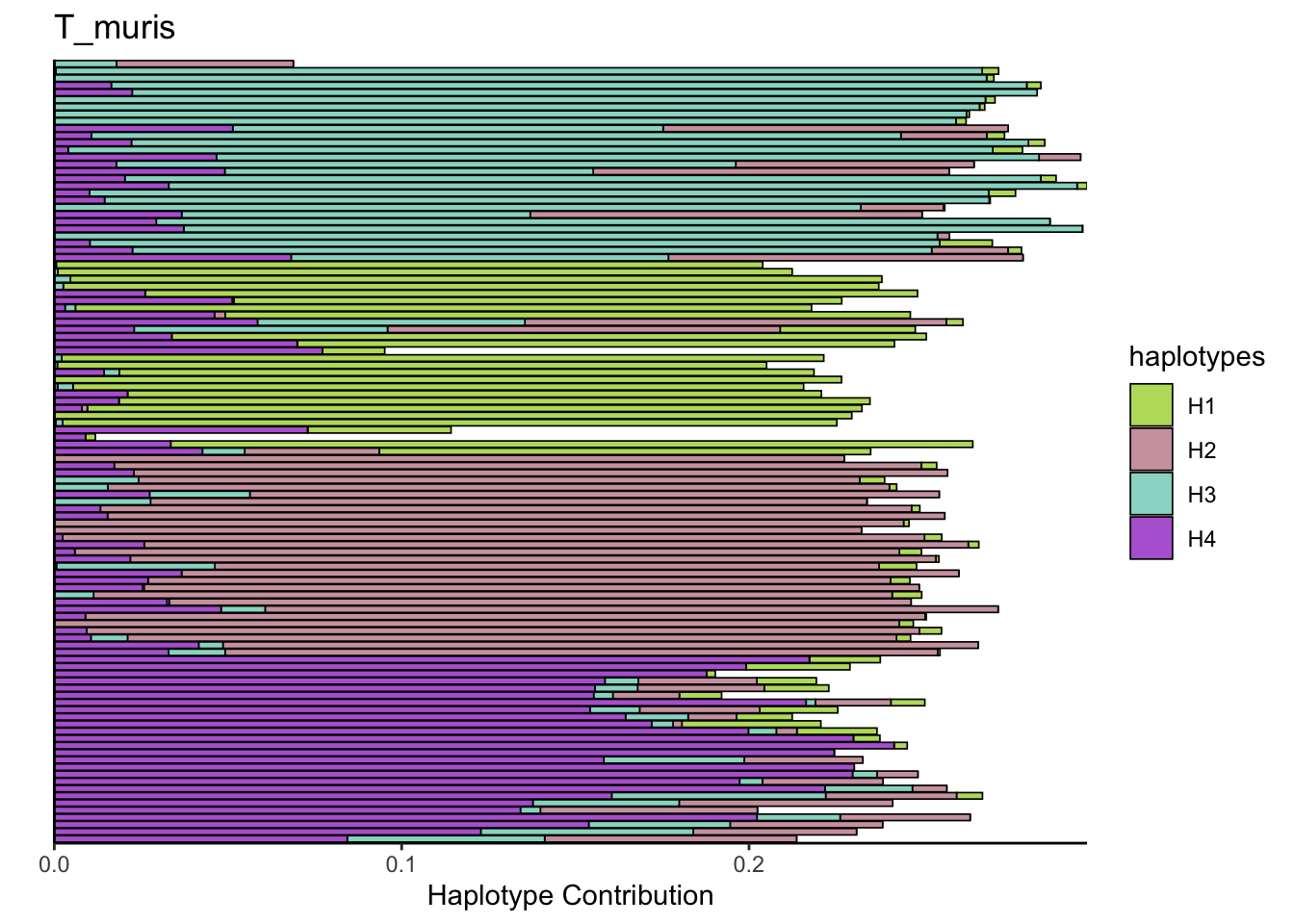
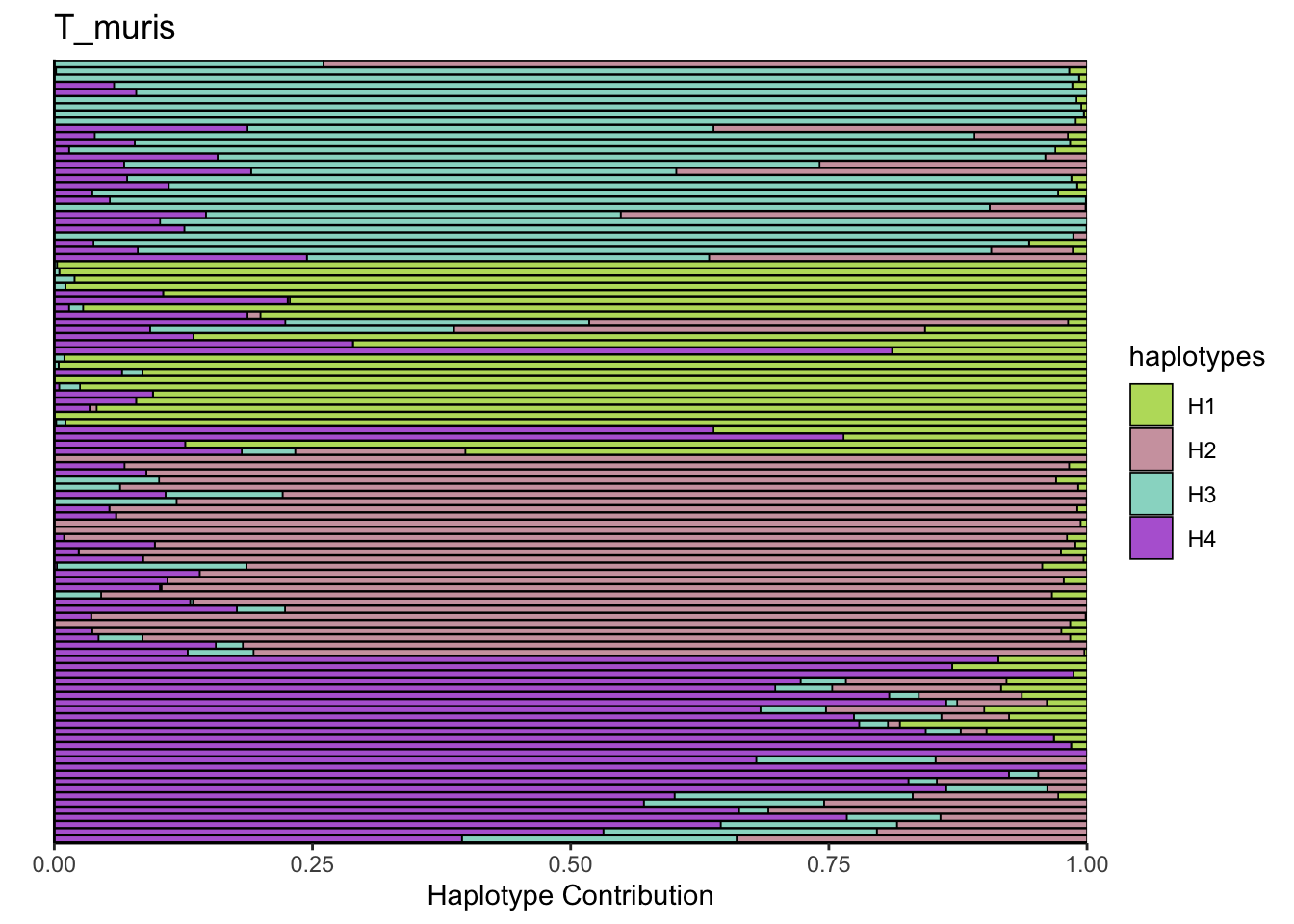
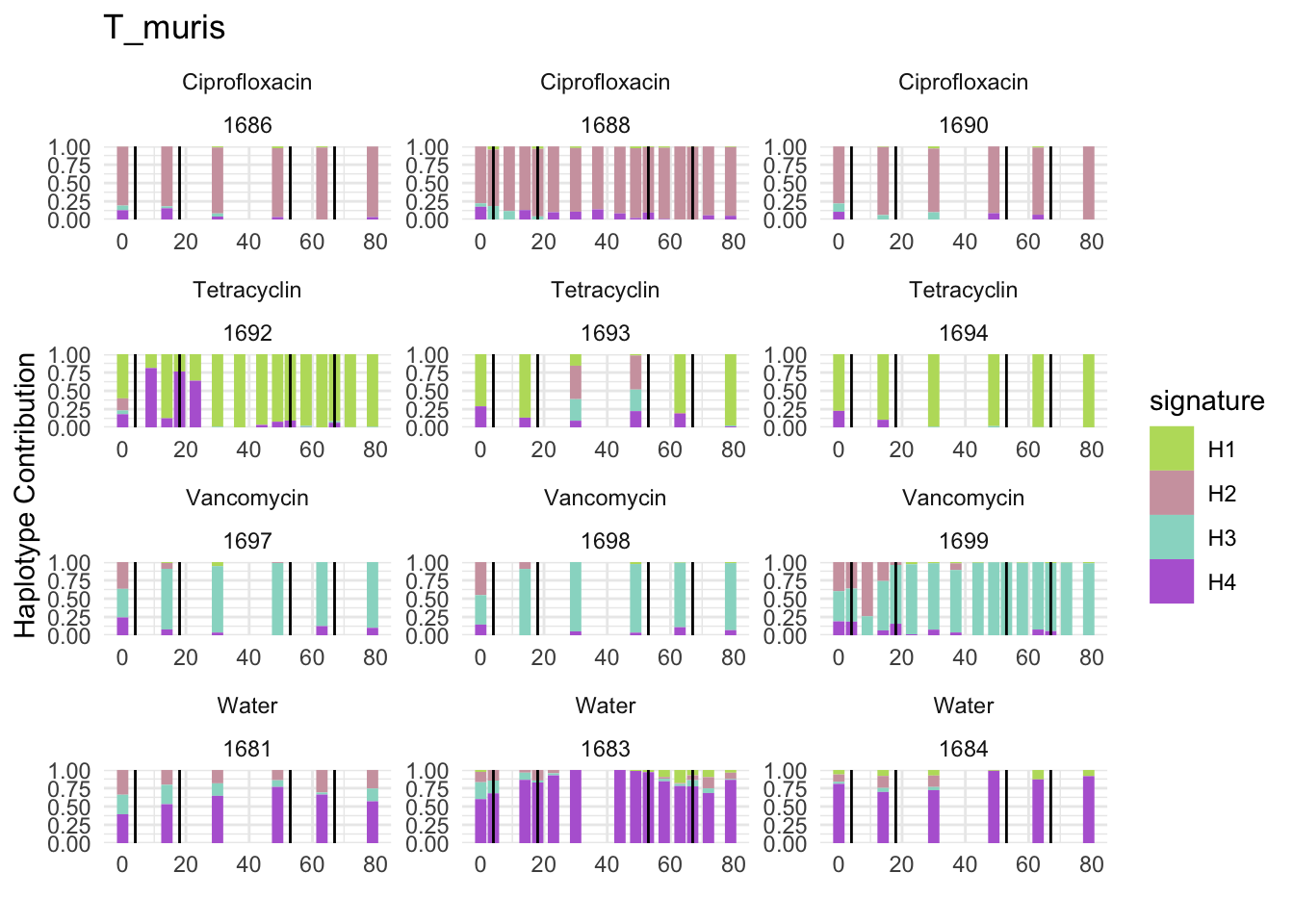
6.4 Blautia_coccoides
## 226# gof <- assessNumberHaplotyes(data.matrix(omm), 2:8, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 3)
plotHaplotypeMap(decomposed)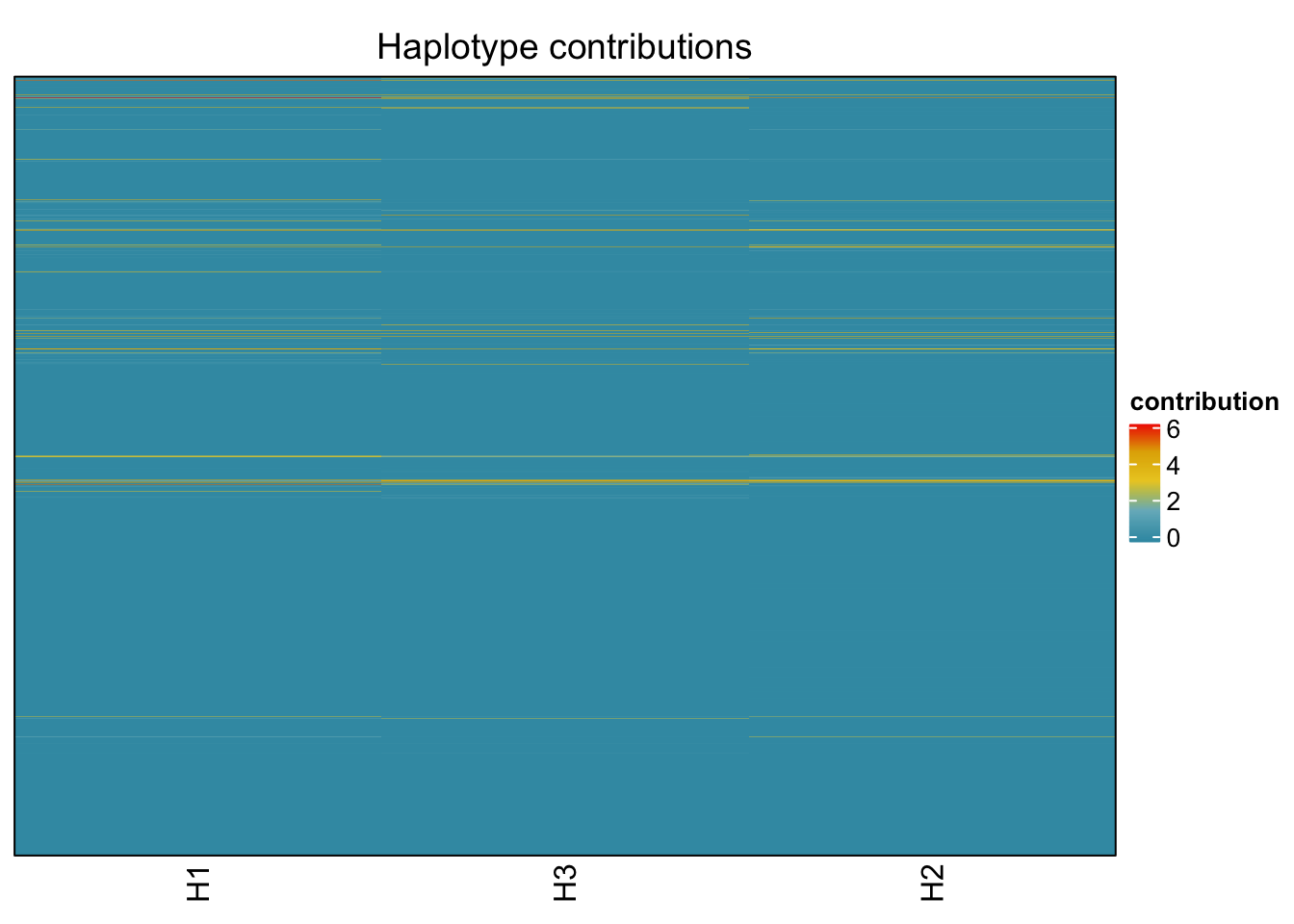
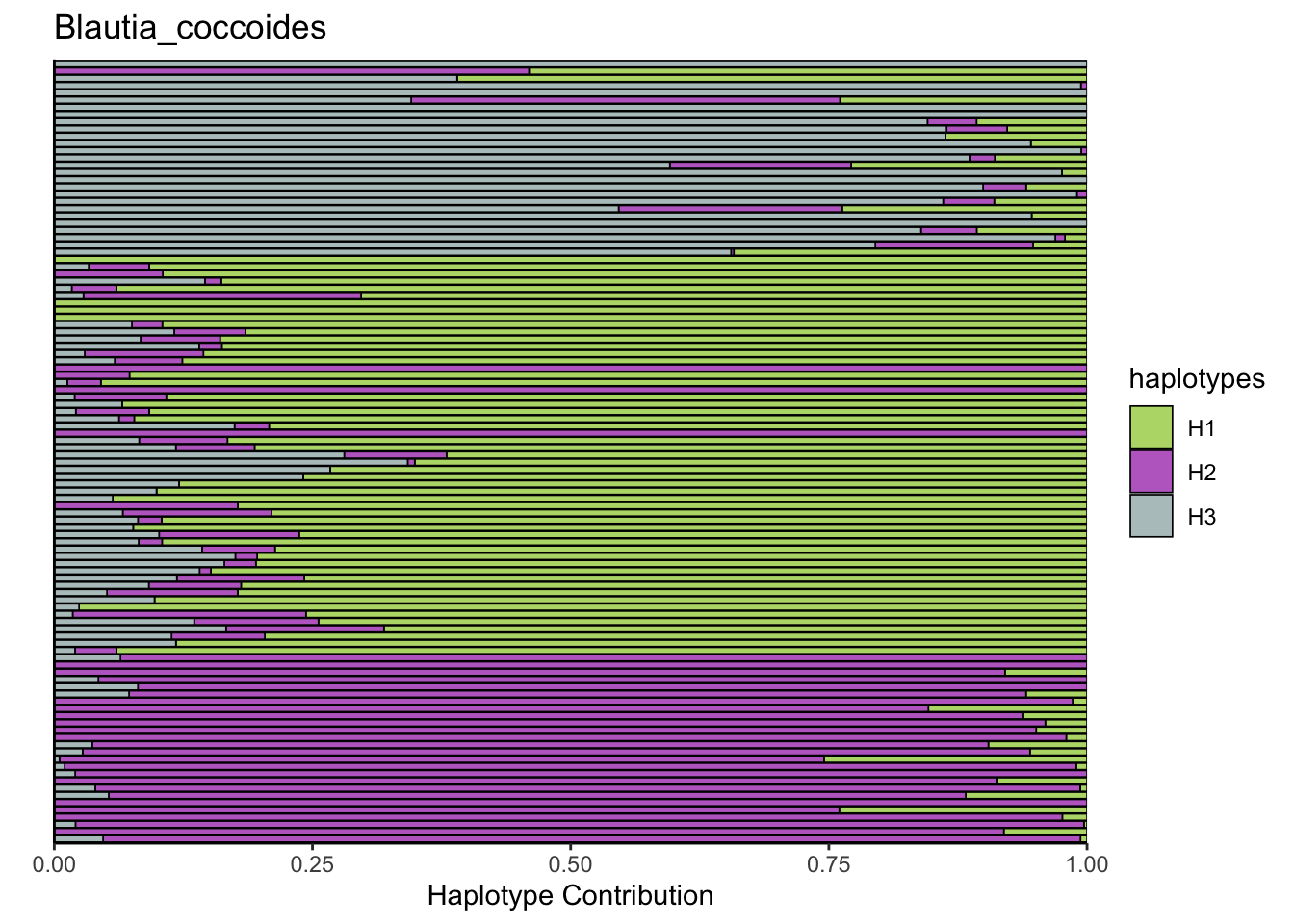
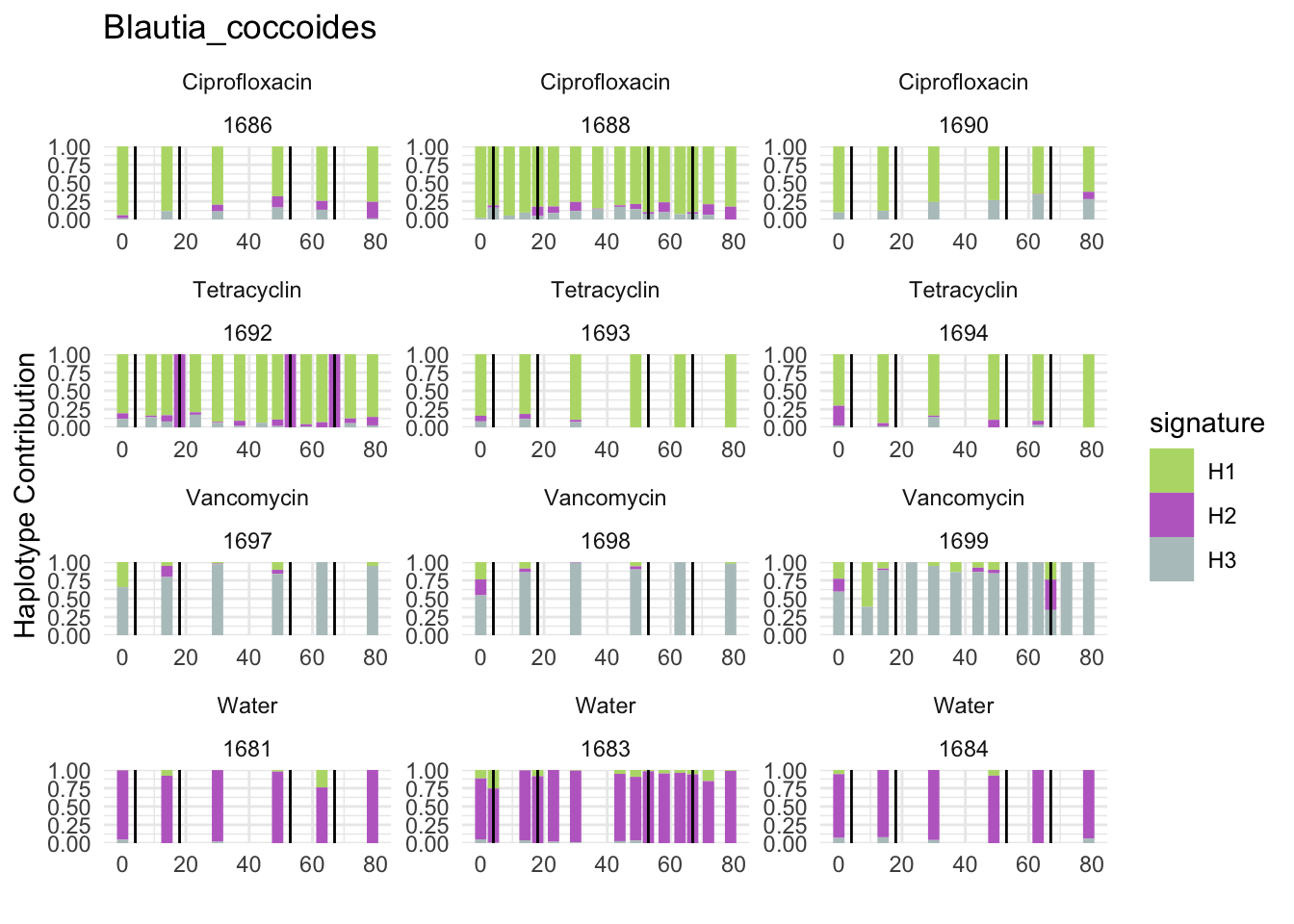
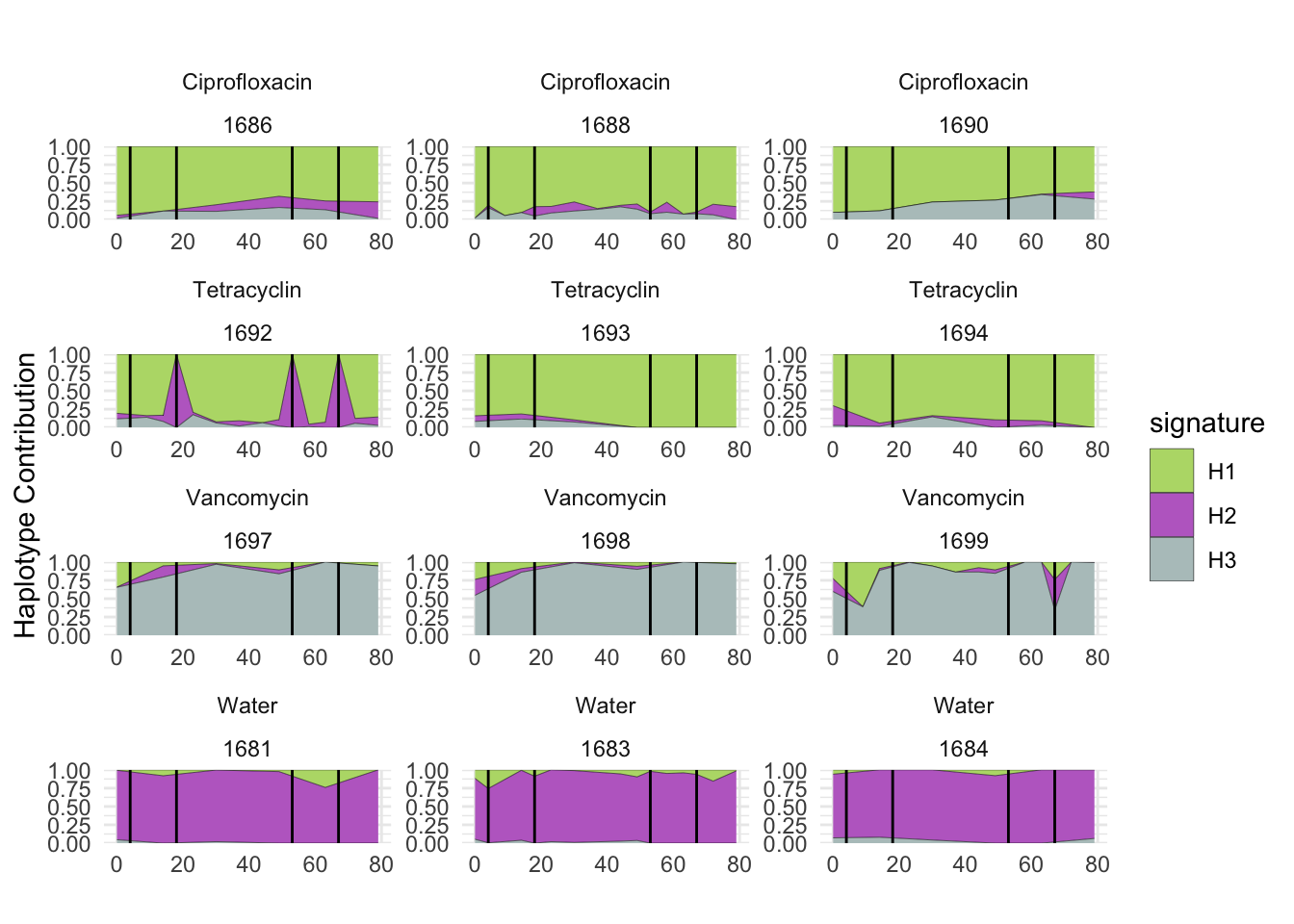
6.5 Akkermansia_muciniphila
bug <- "Akkermansia_muciniphila"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 6)
plotHaplotypeMap(decomposed)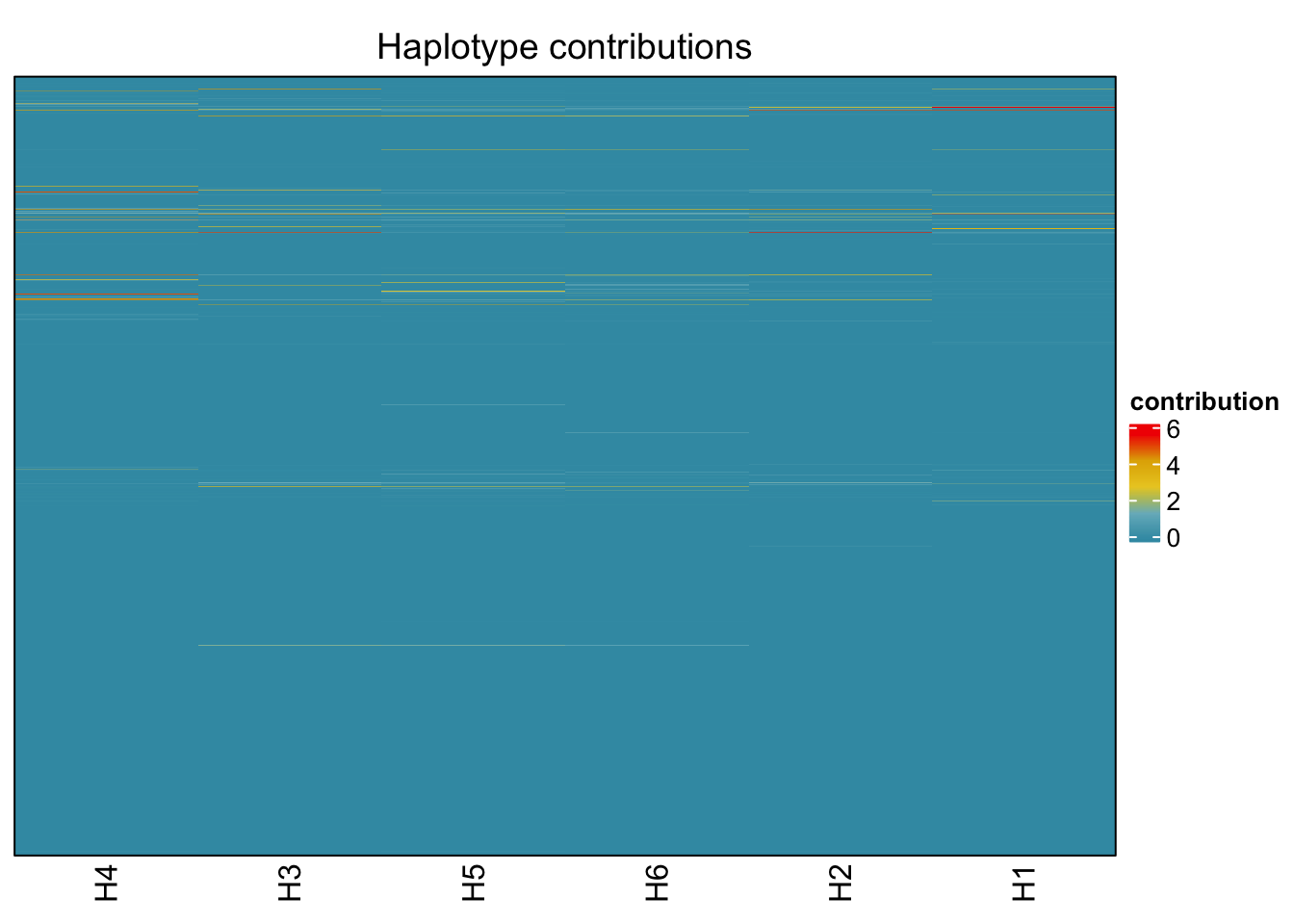
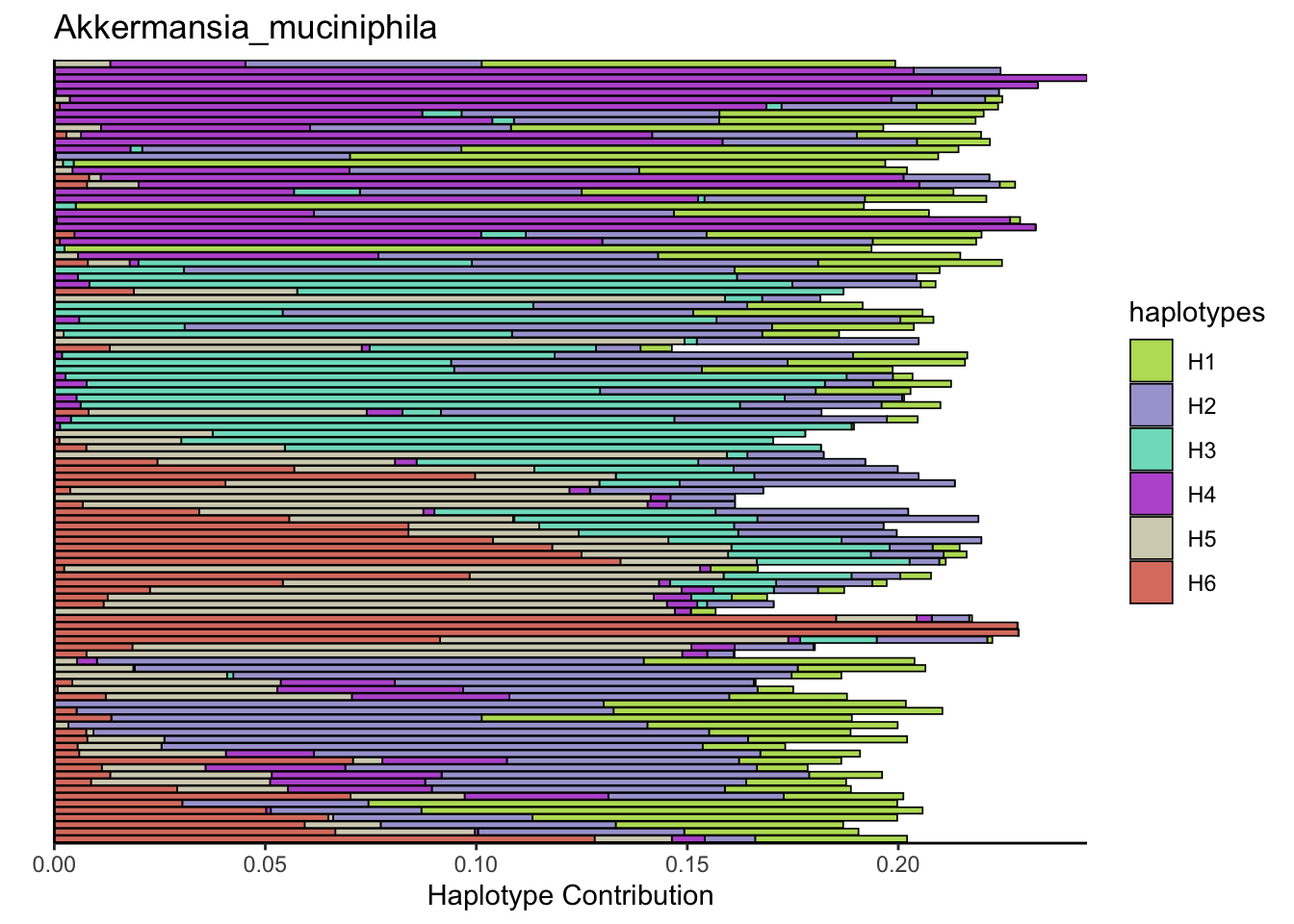
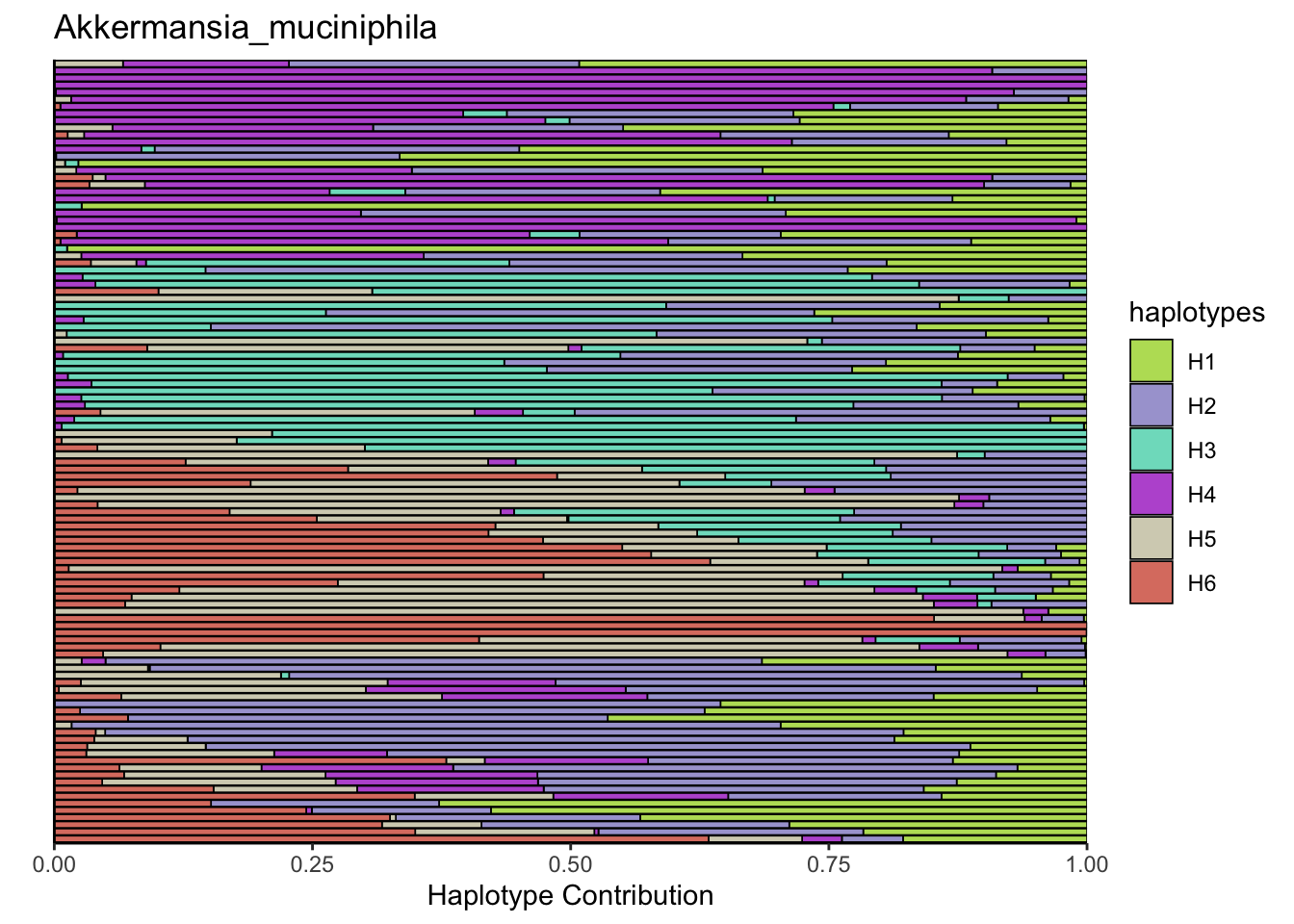
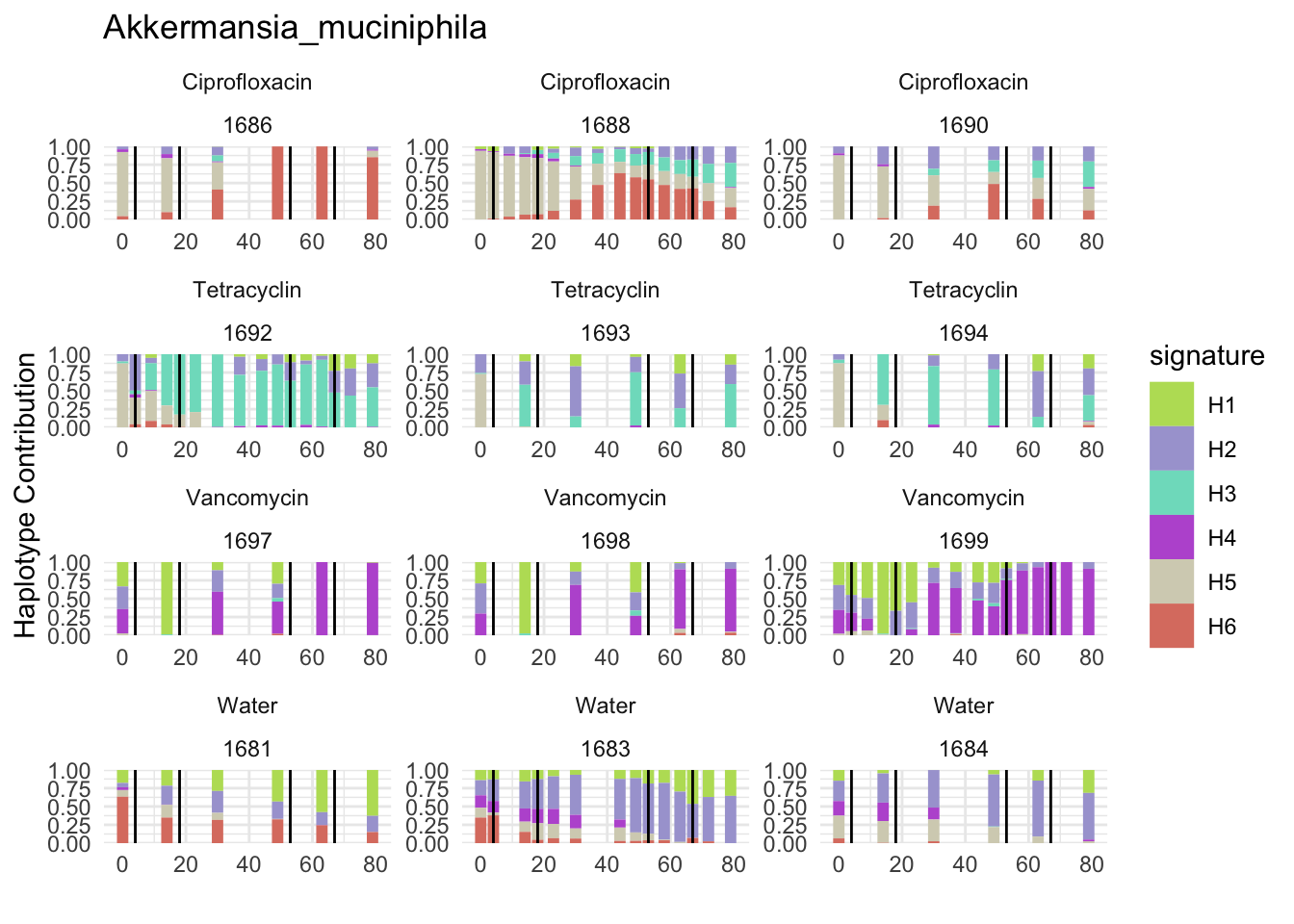
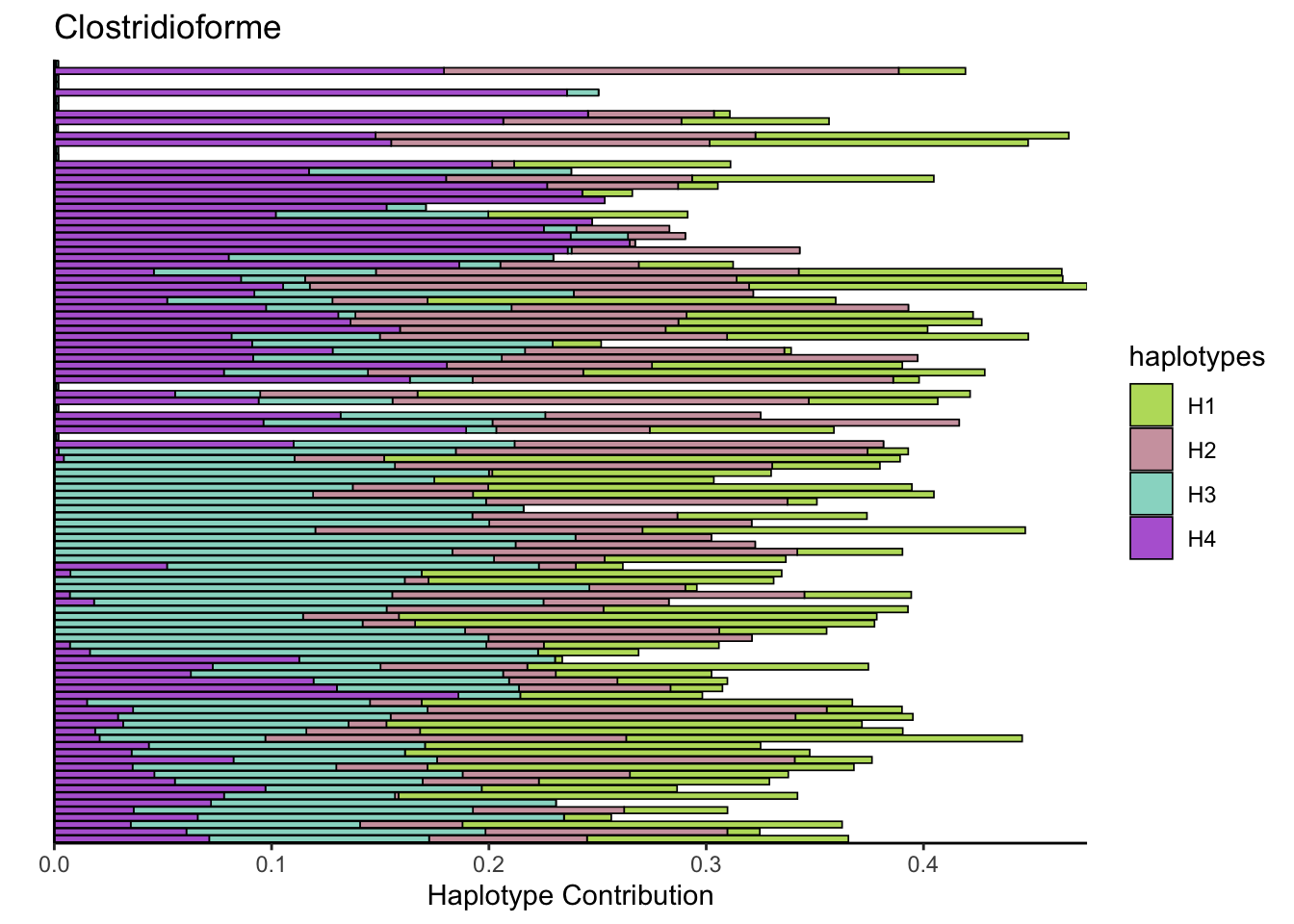
6.6 Clostridioforme
bug <- "Clostridioforme"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 4)
plotHaplotypeMap(decomposed)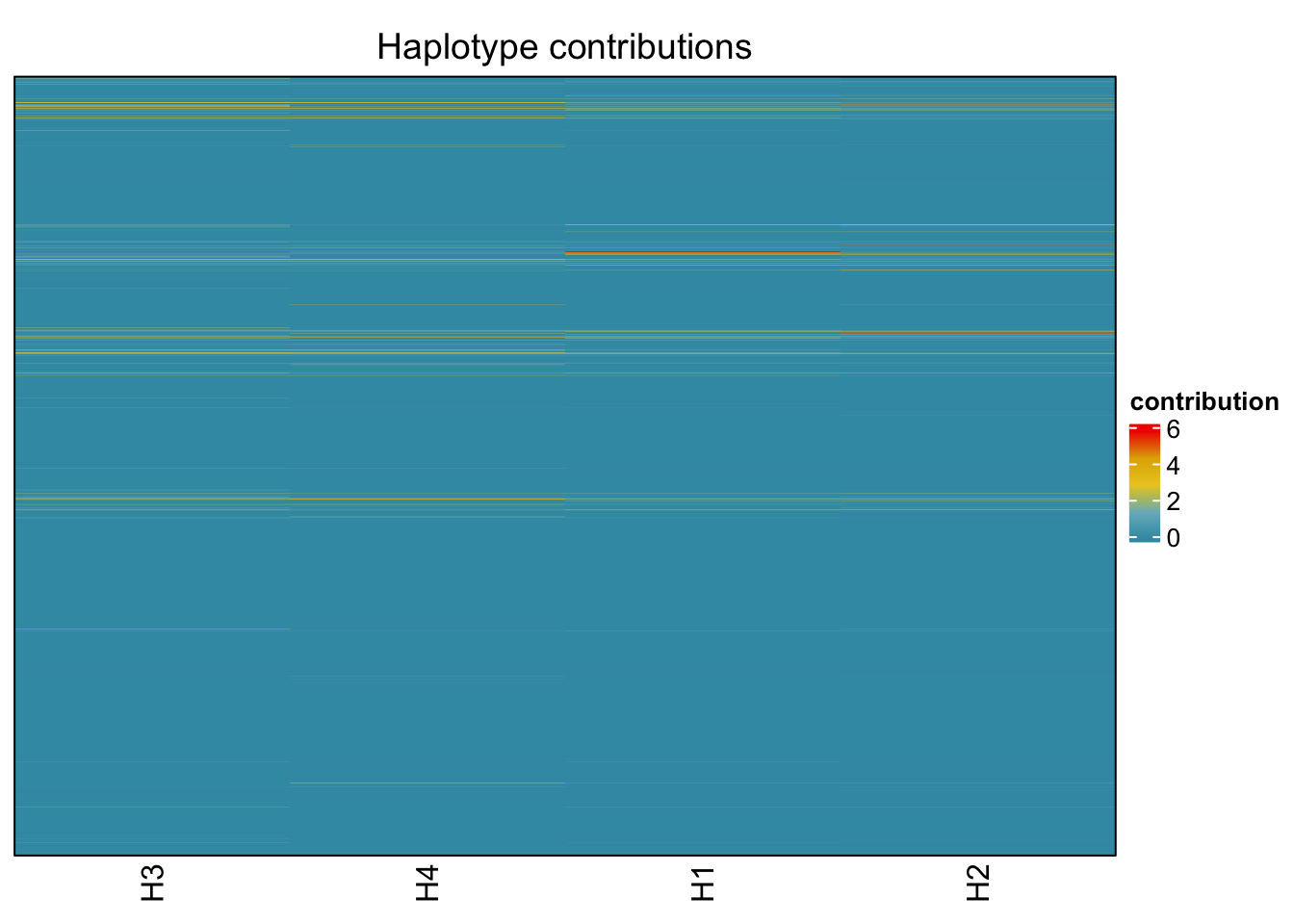
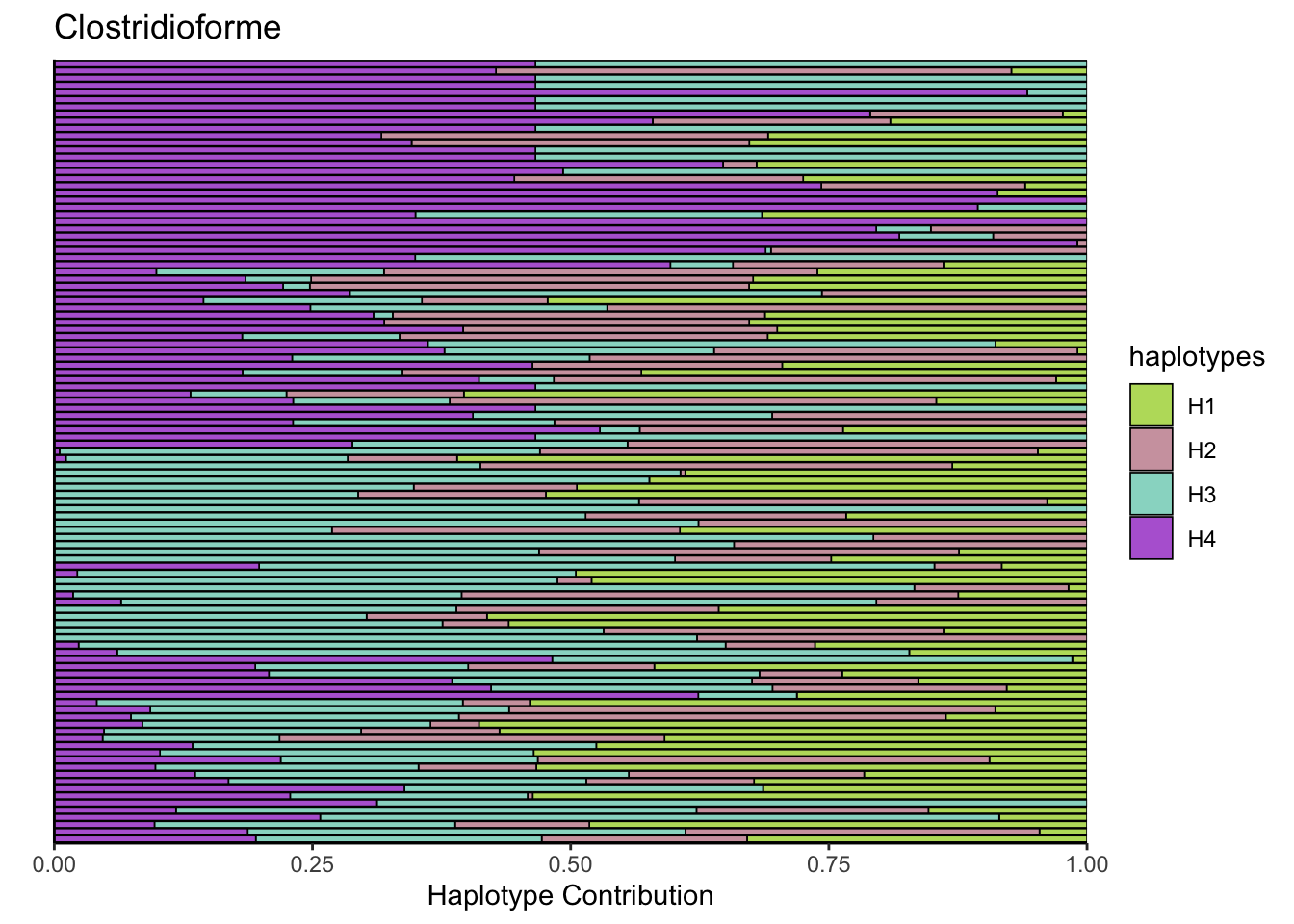
6.7 Muribaculum_intestinale
bug <- "Muribaculum_intestinale"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 6)
plotHaplotypeMap(decomposed)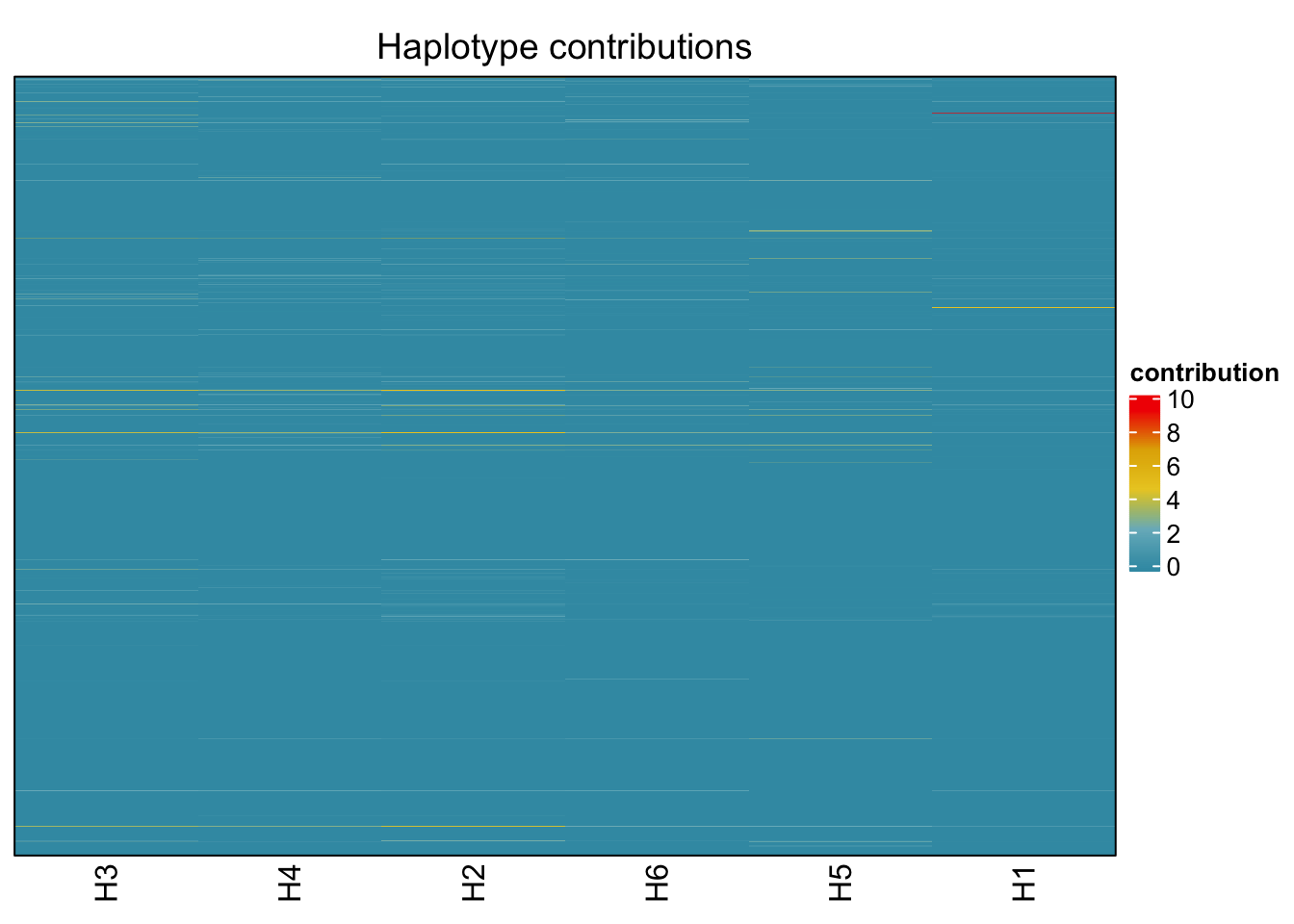
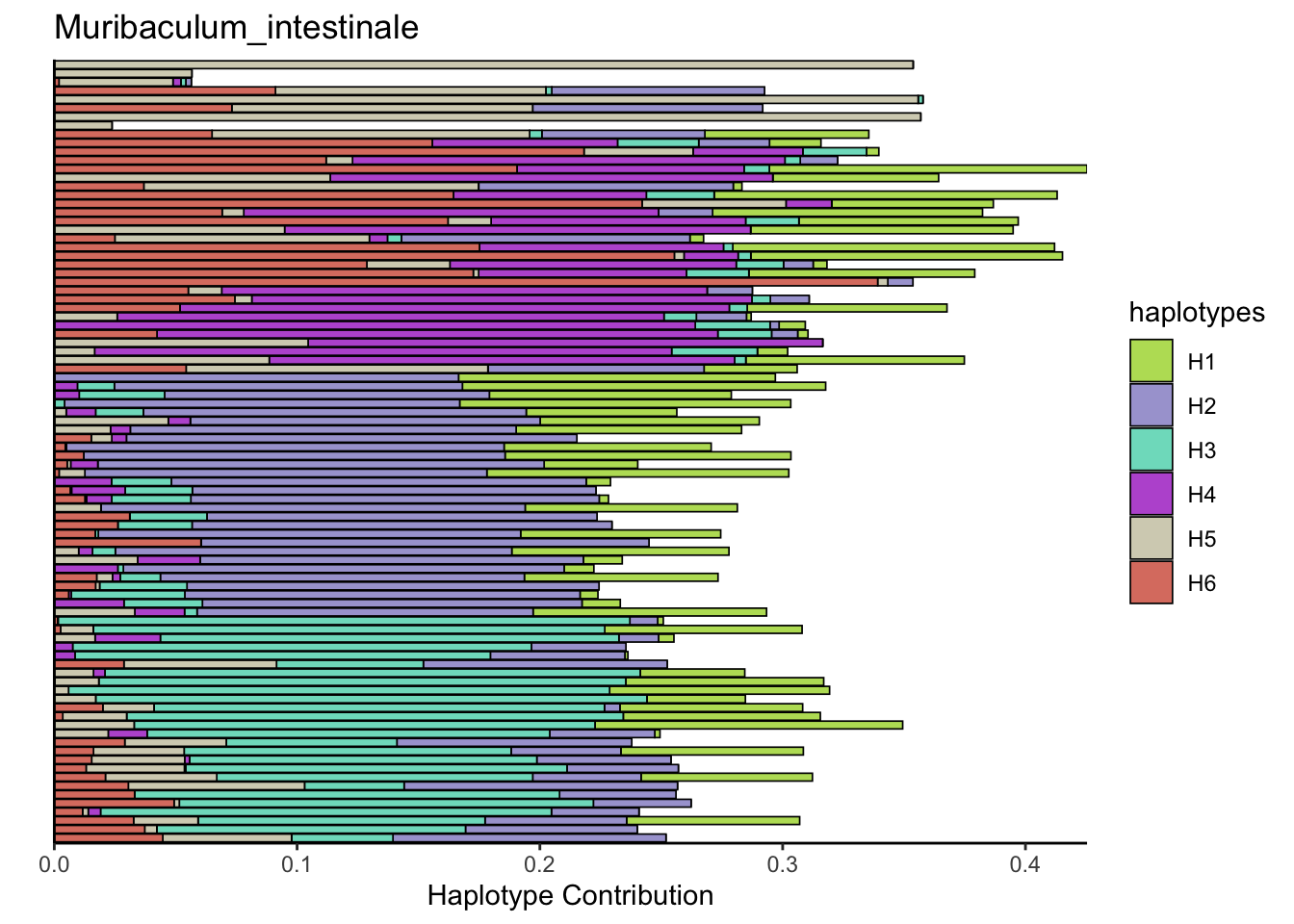
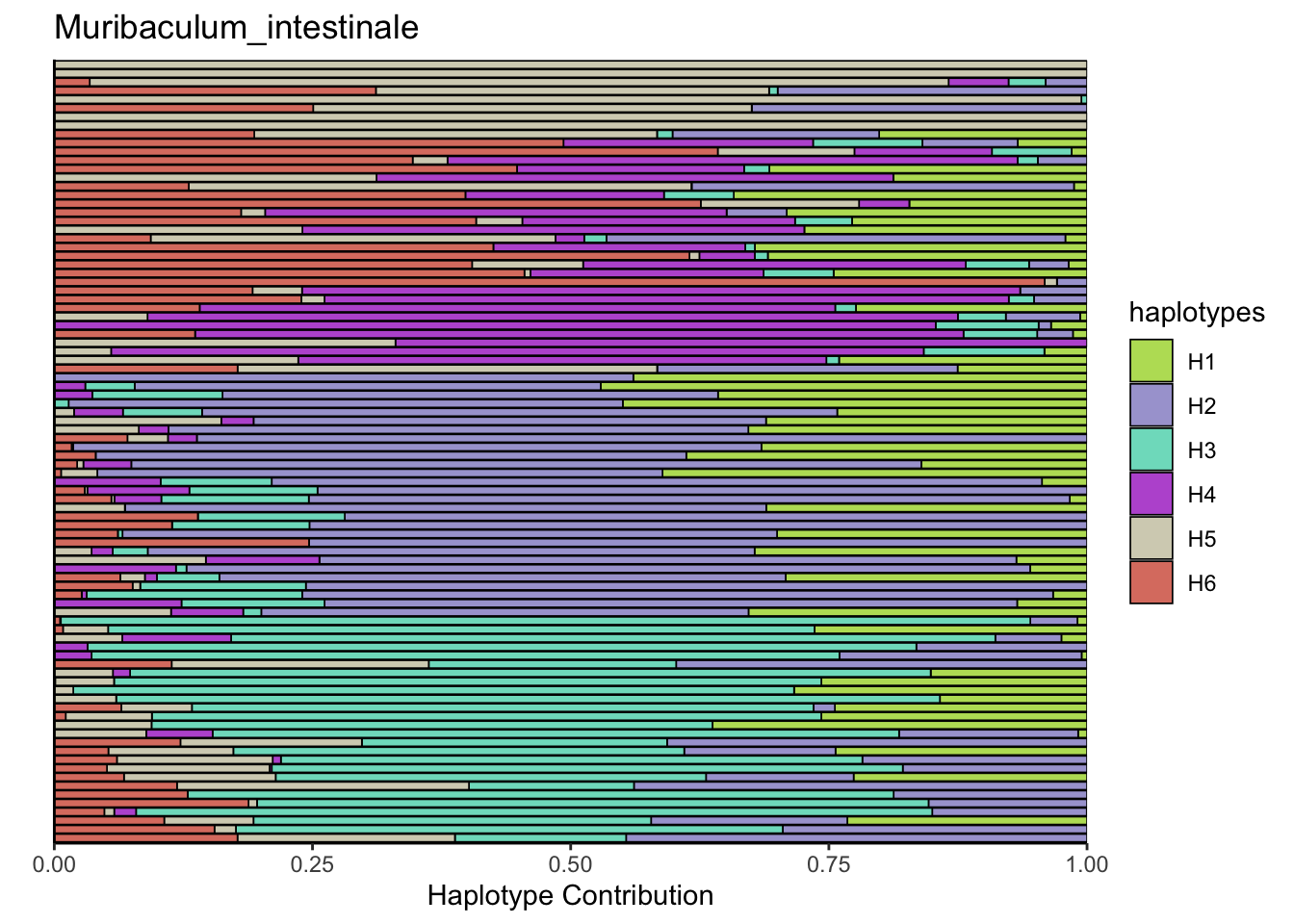
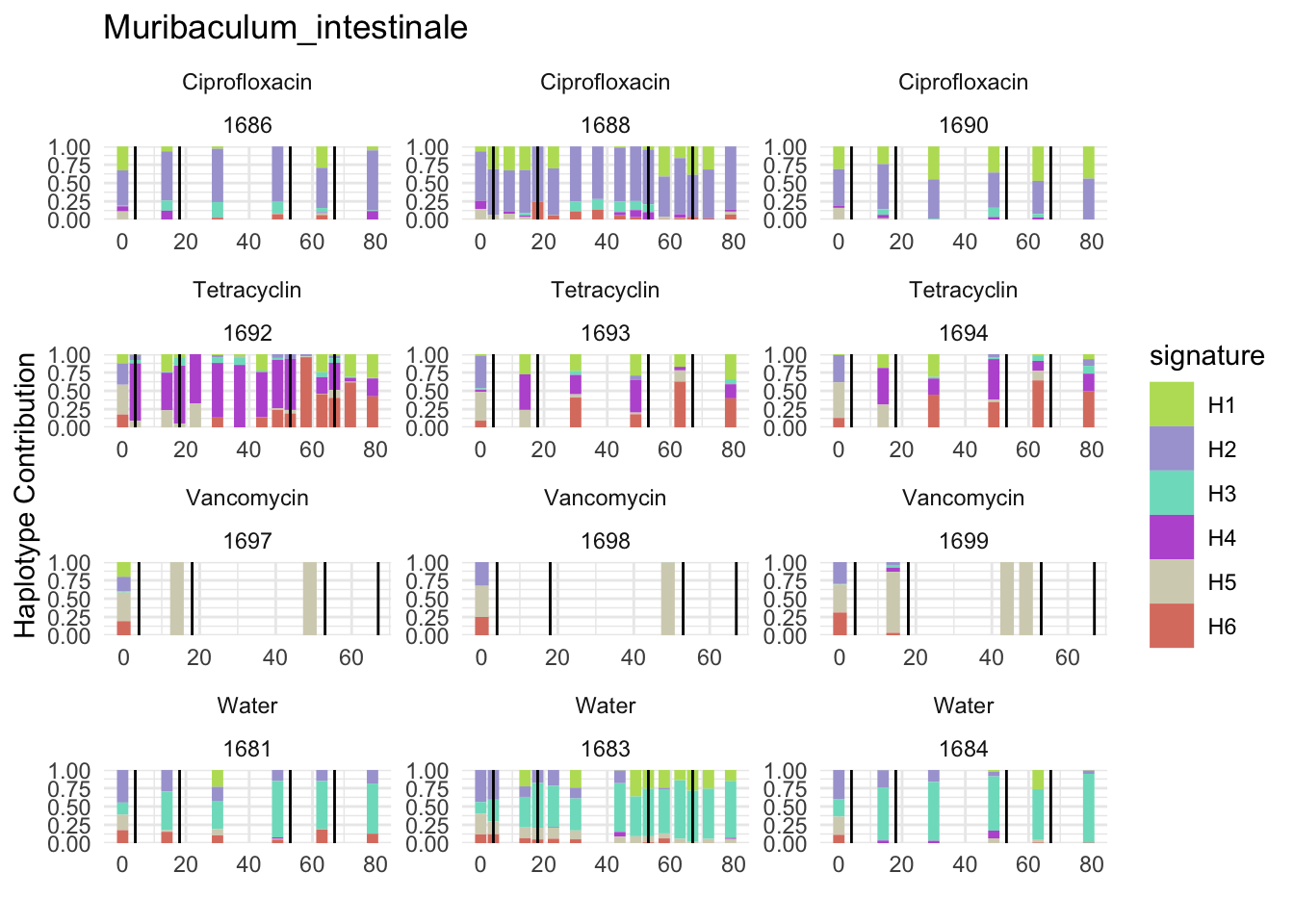
6.8 T_muris
bug <- "T_muris"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 4)
plotHaplotypeMap(decomposed)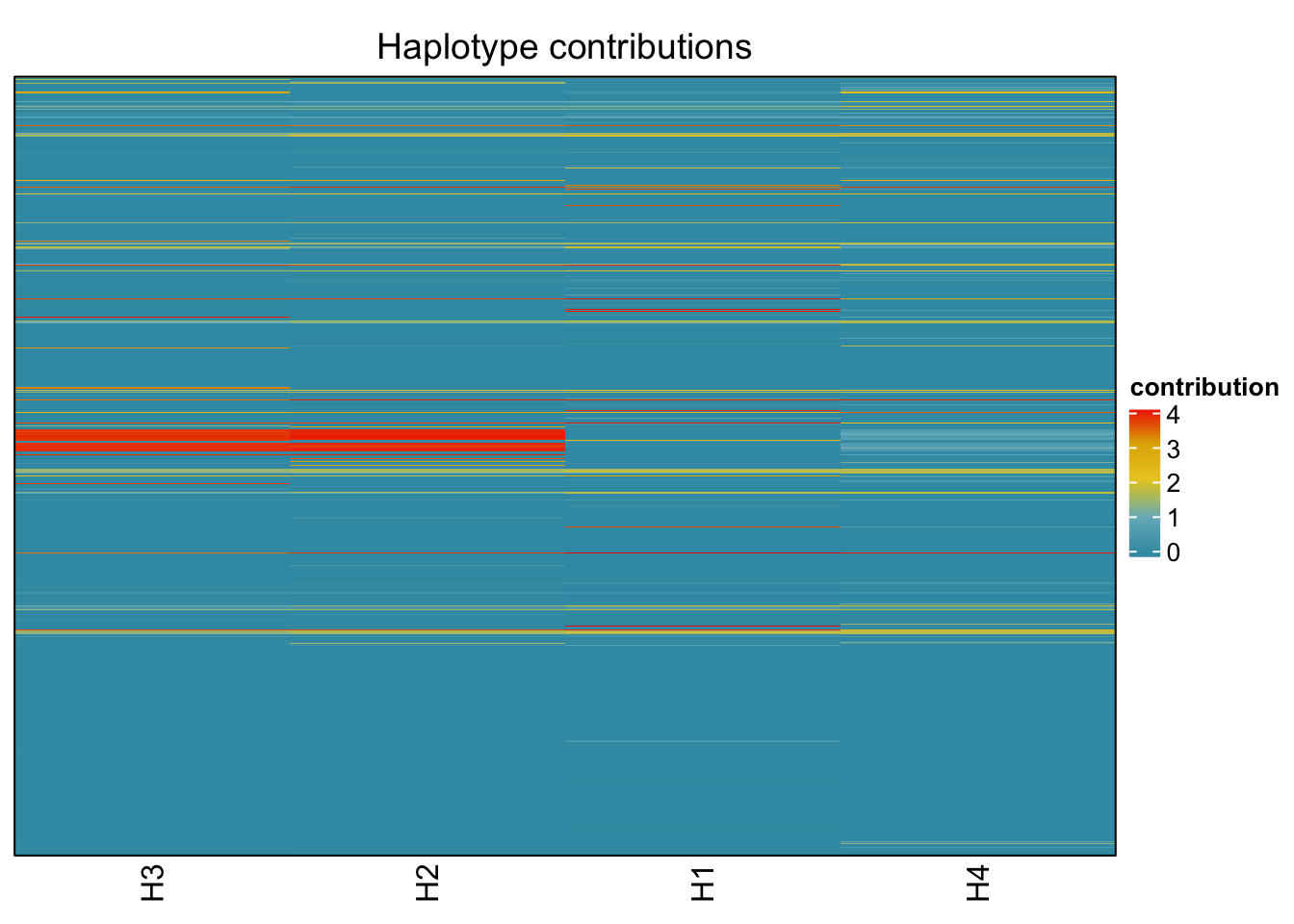
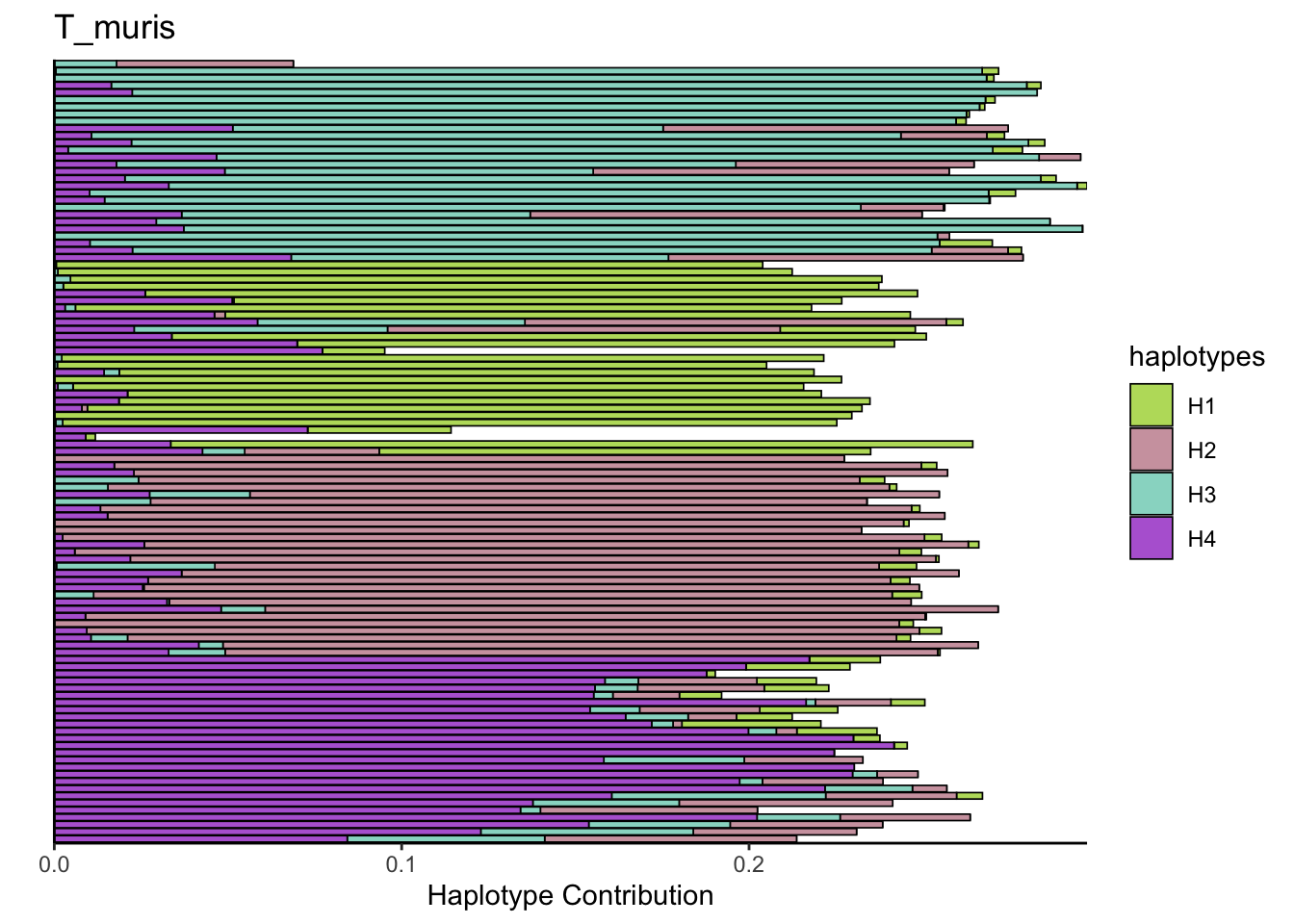
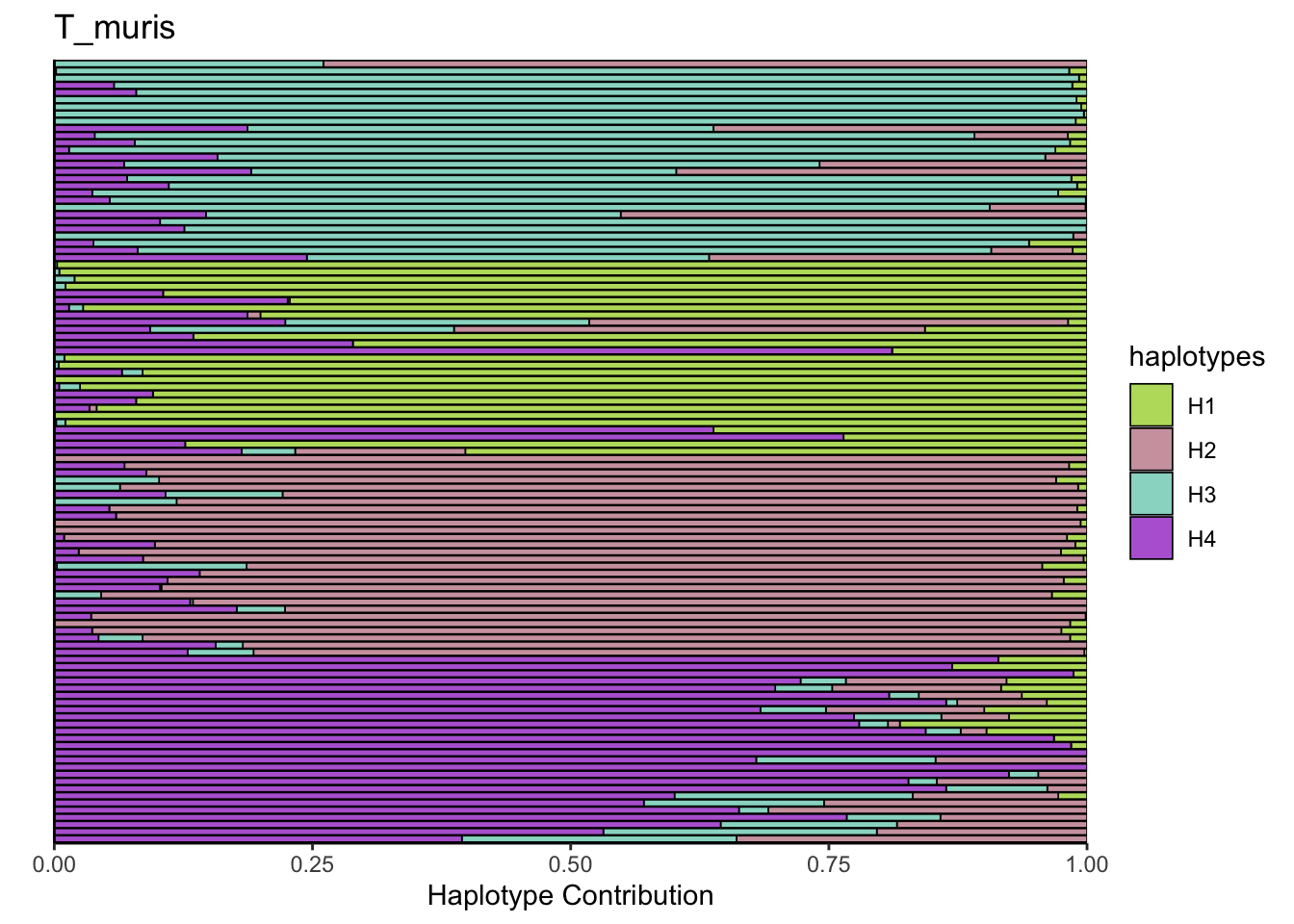
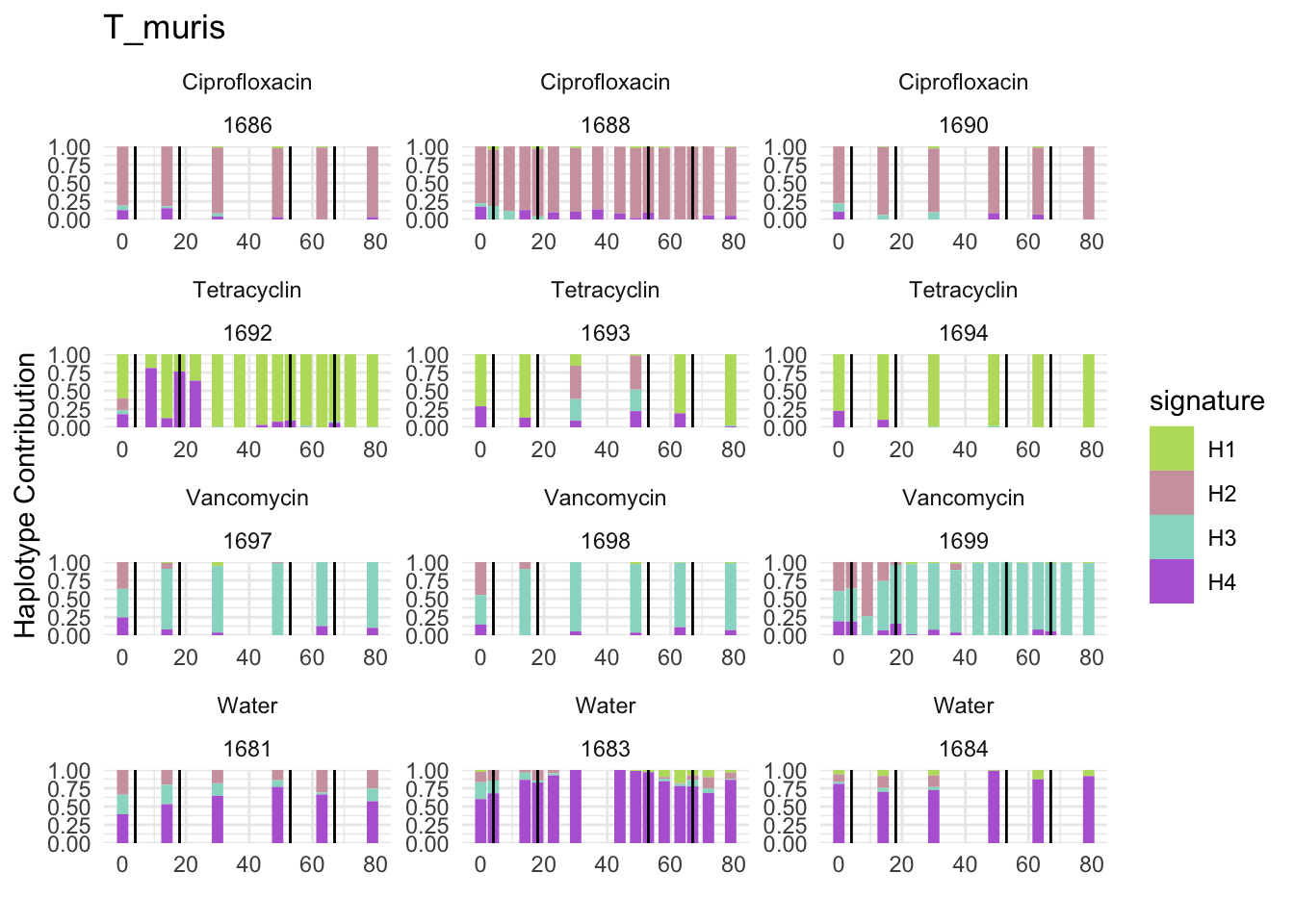
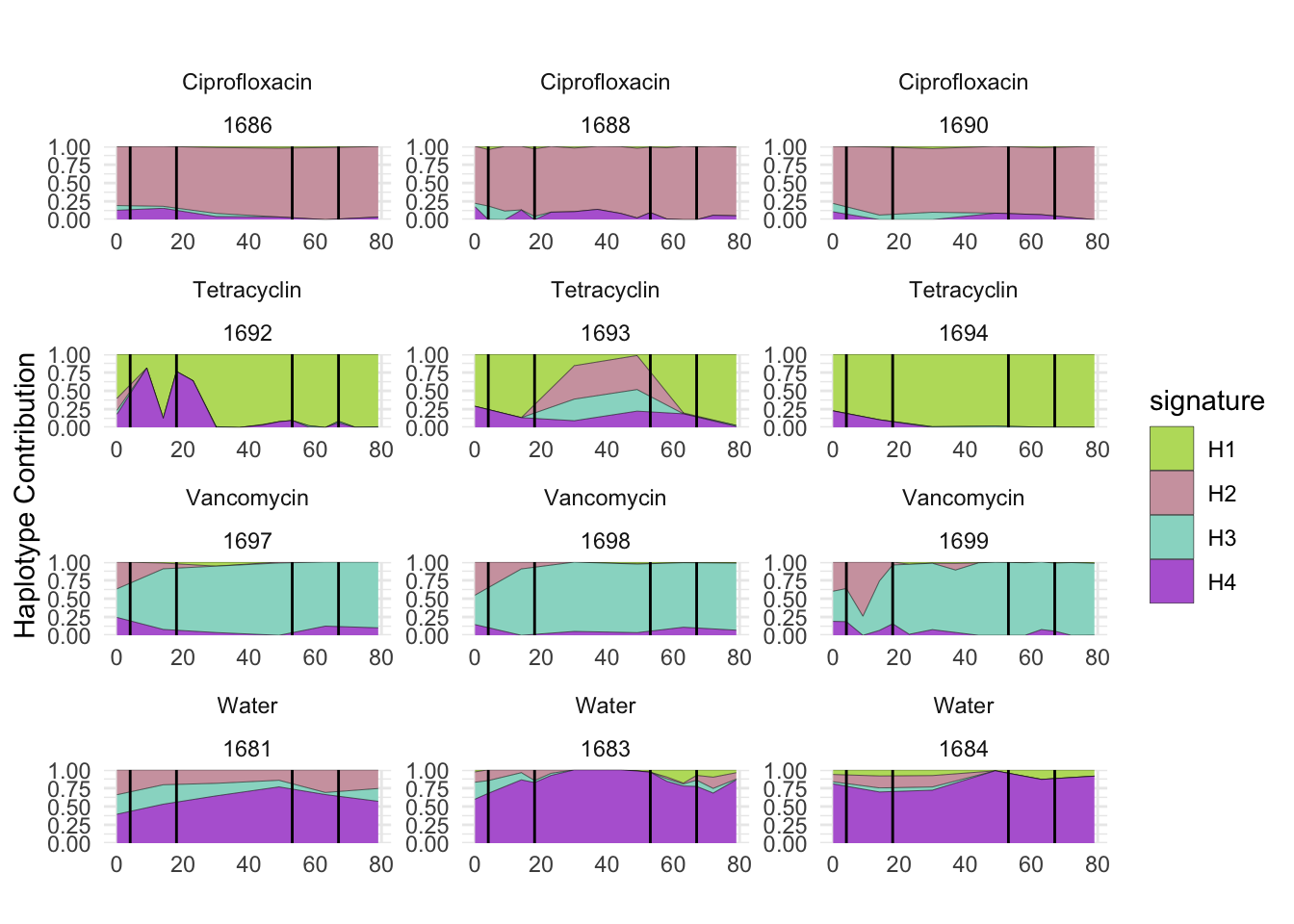
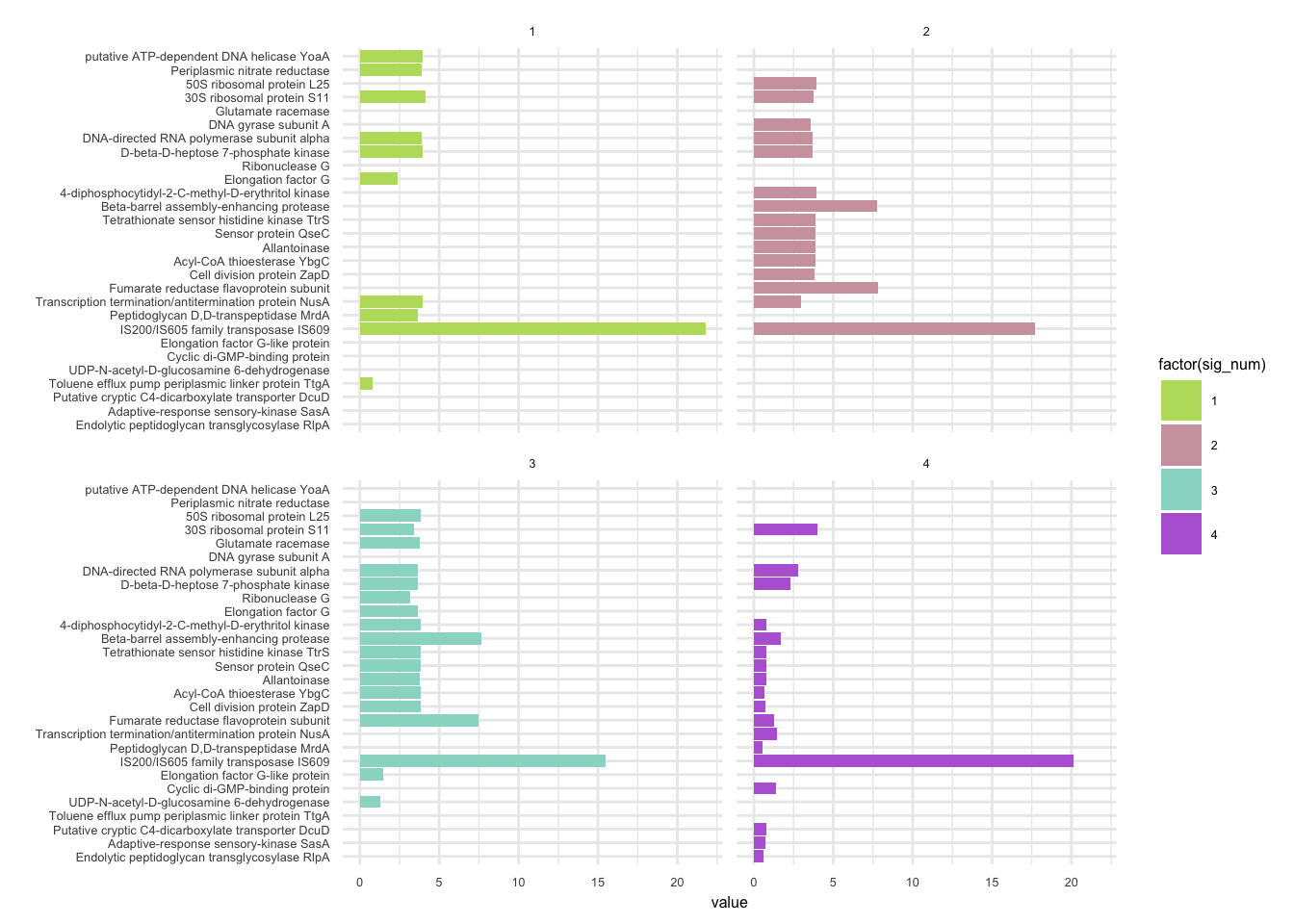
6.9 F_plautii_1
bug <- "F_plautii_1"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 4)
plotHaplotypeMap(decomposed)
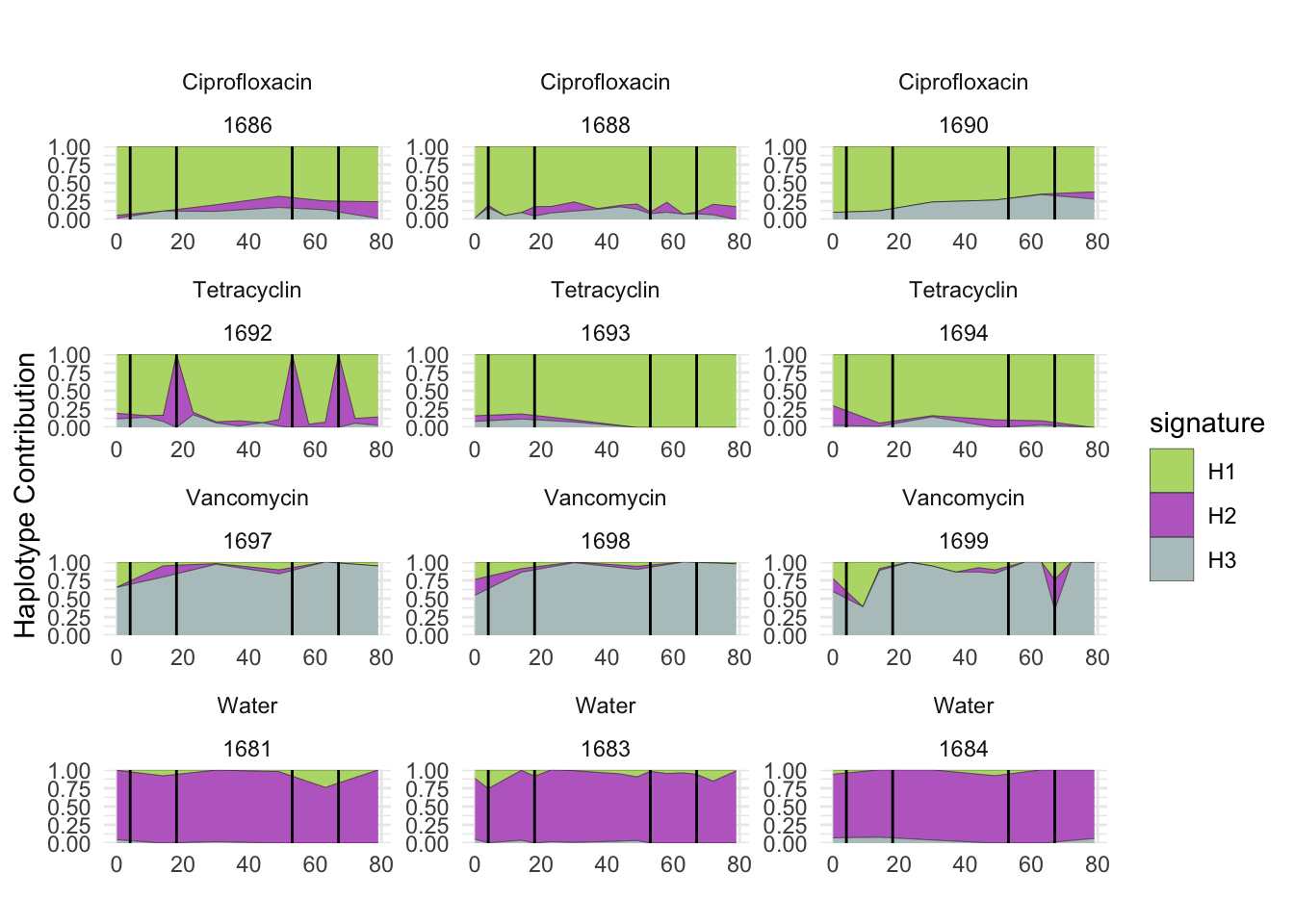
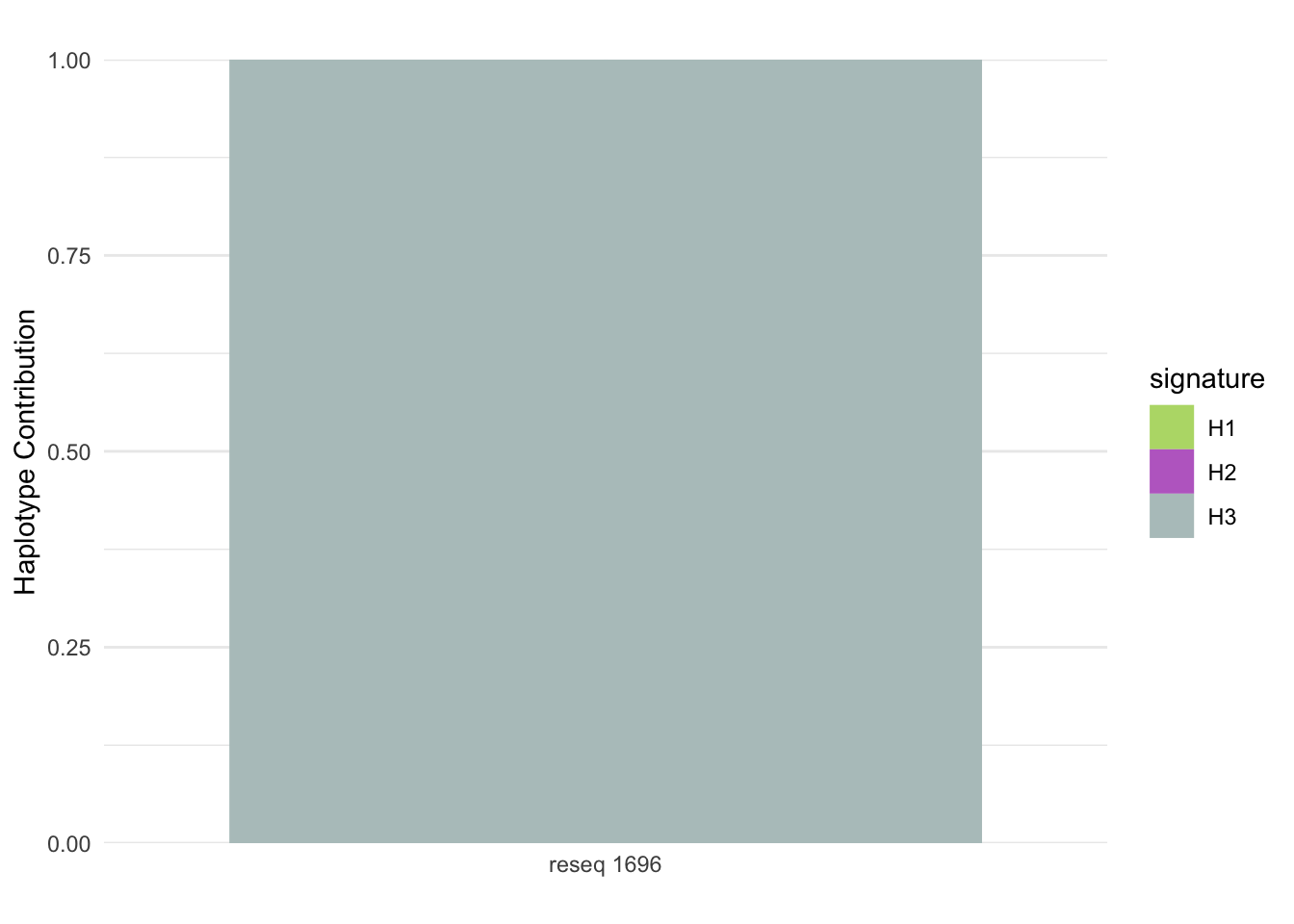
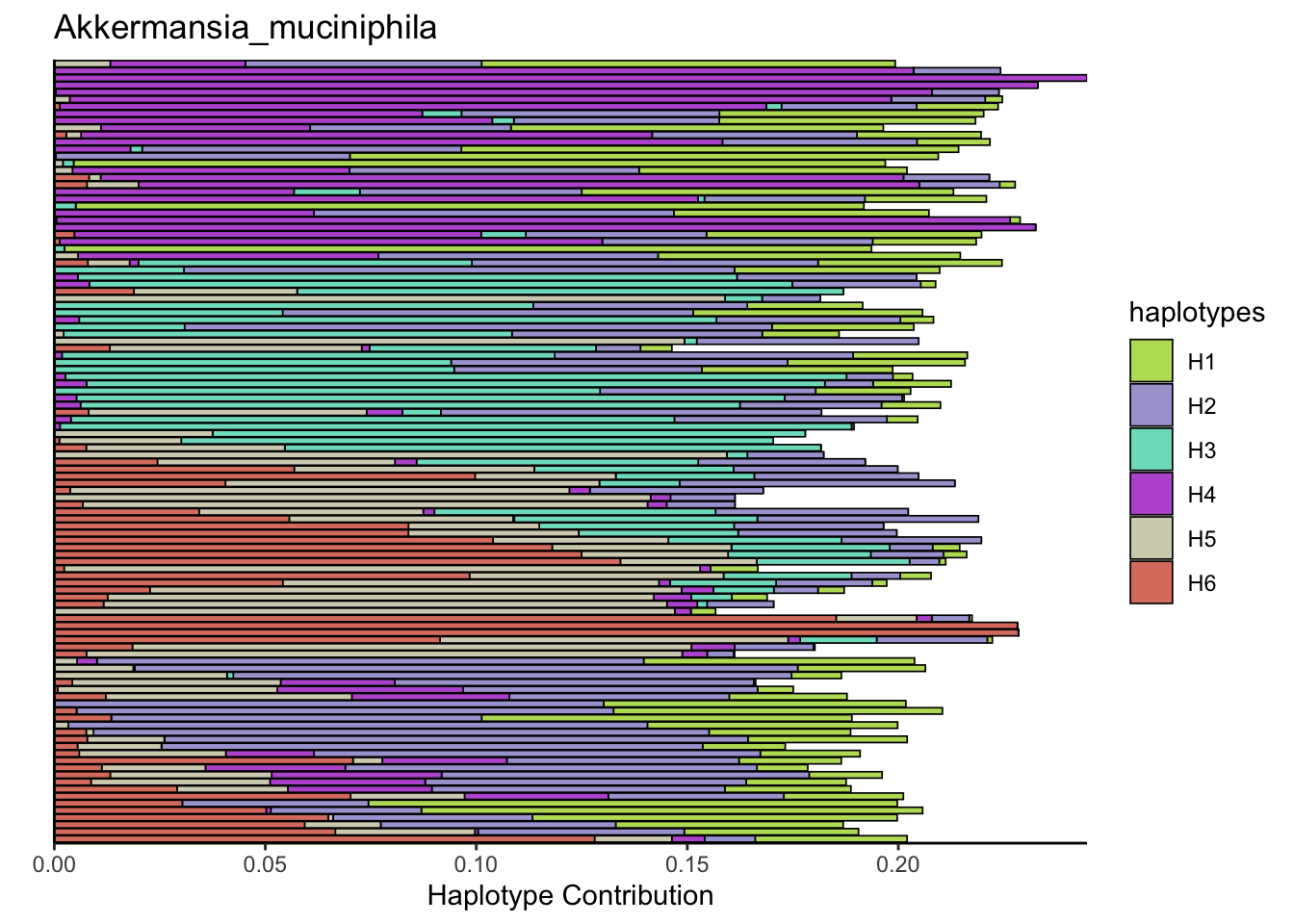
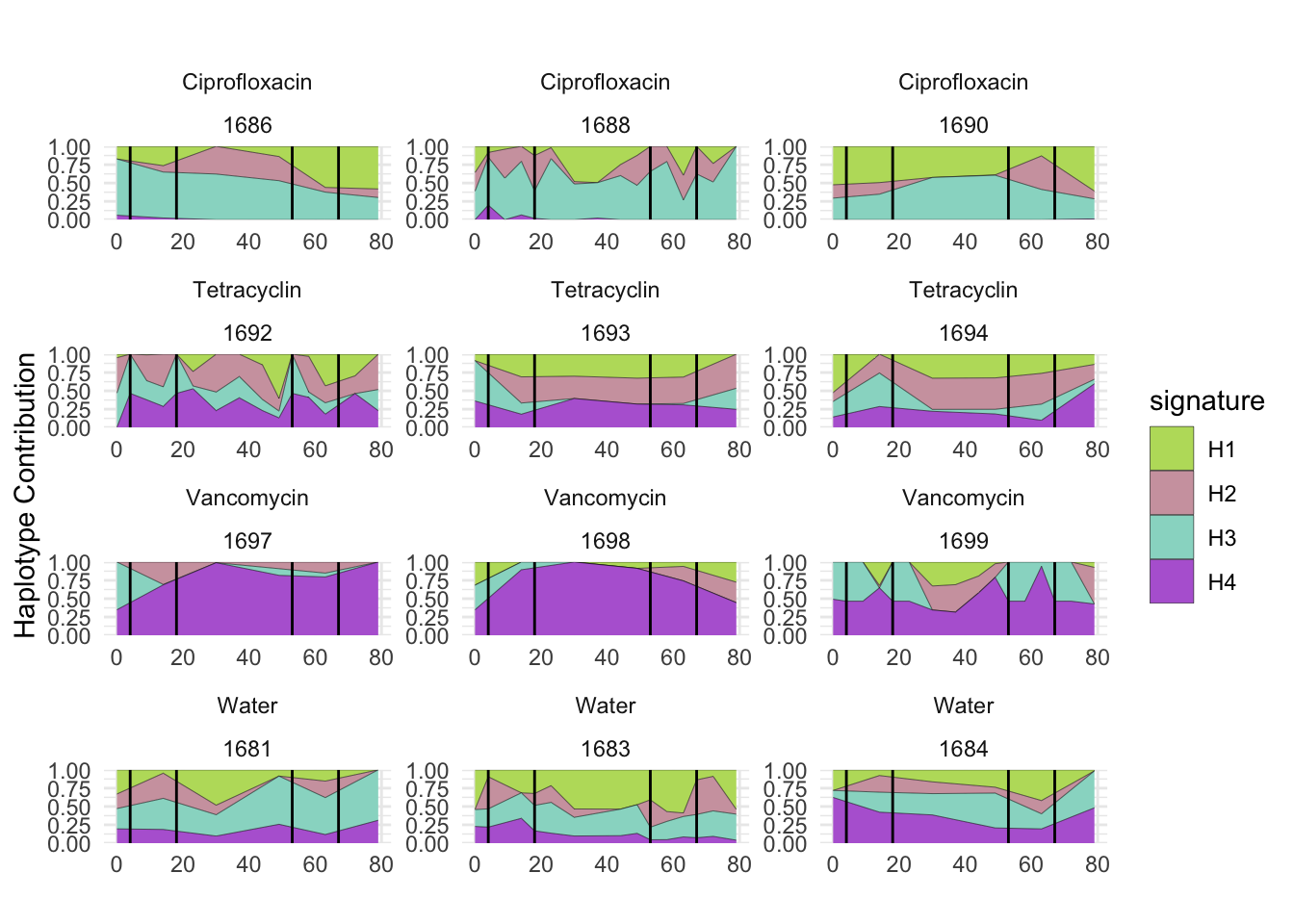
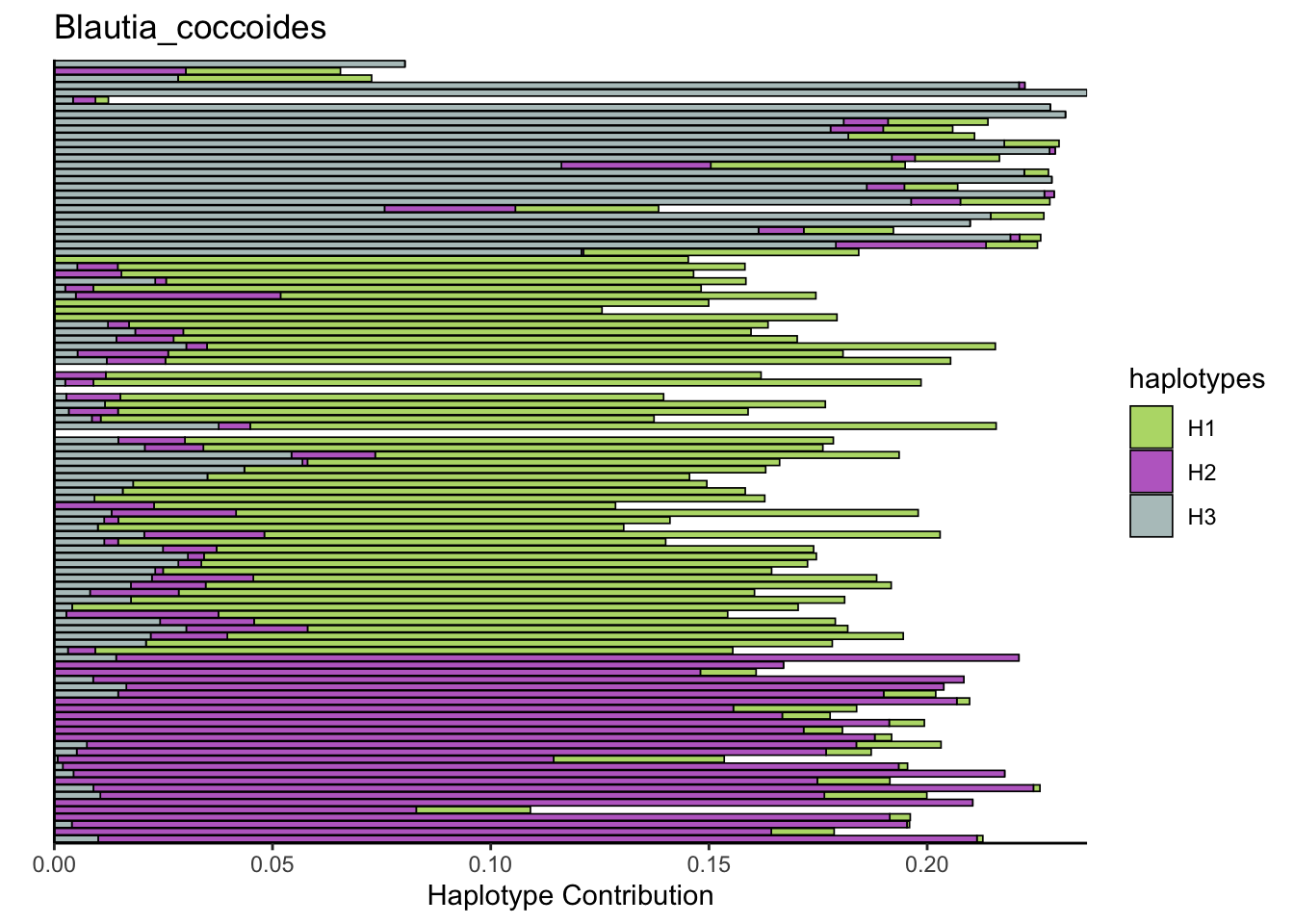
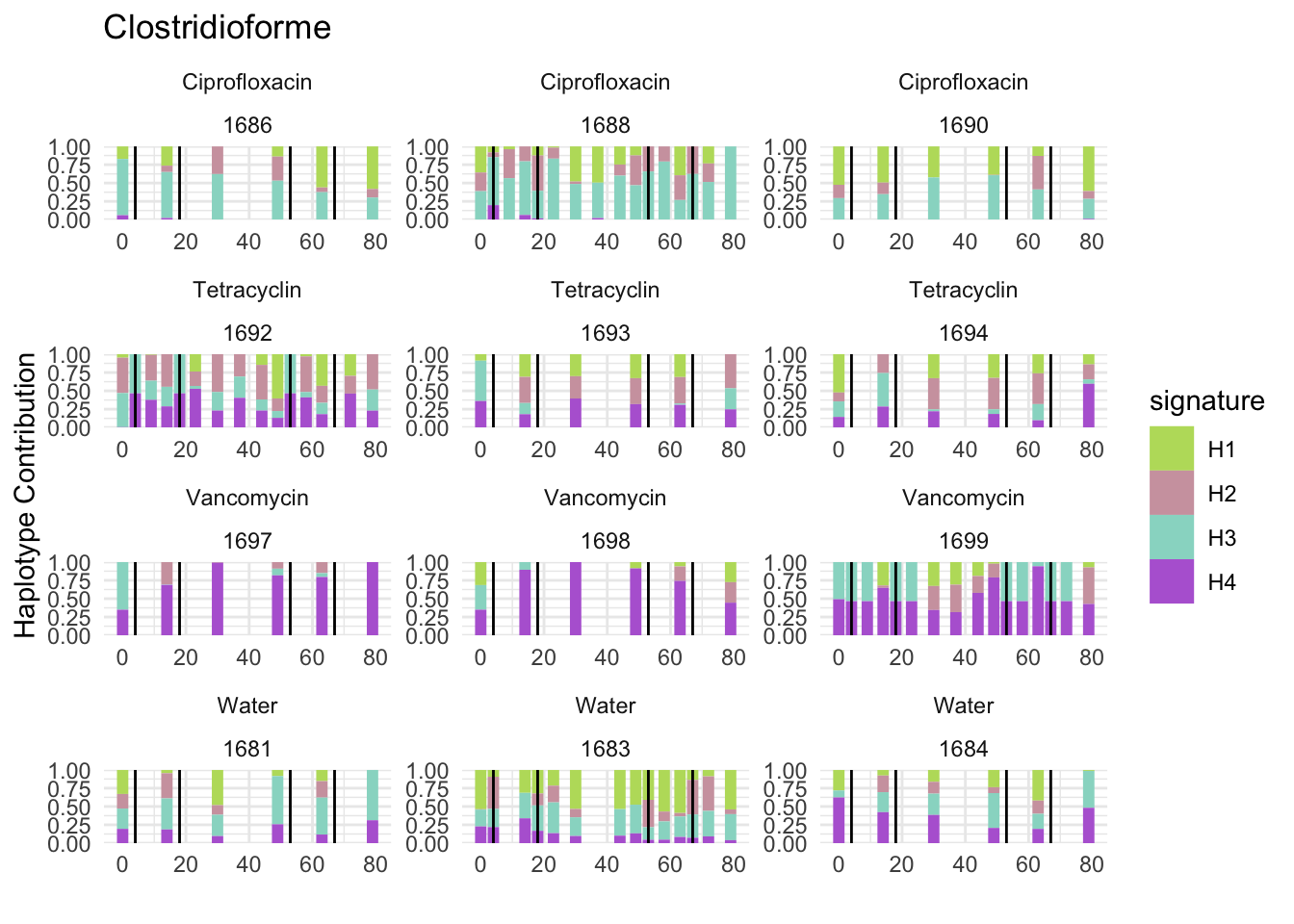
plotSamples(decomposed, normalize = F, remove.sample.names = T, title = bug)
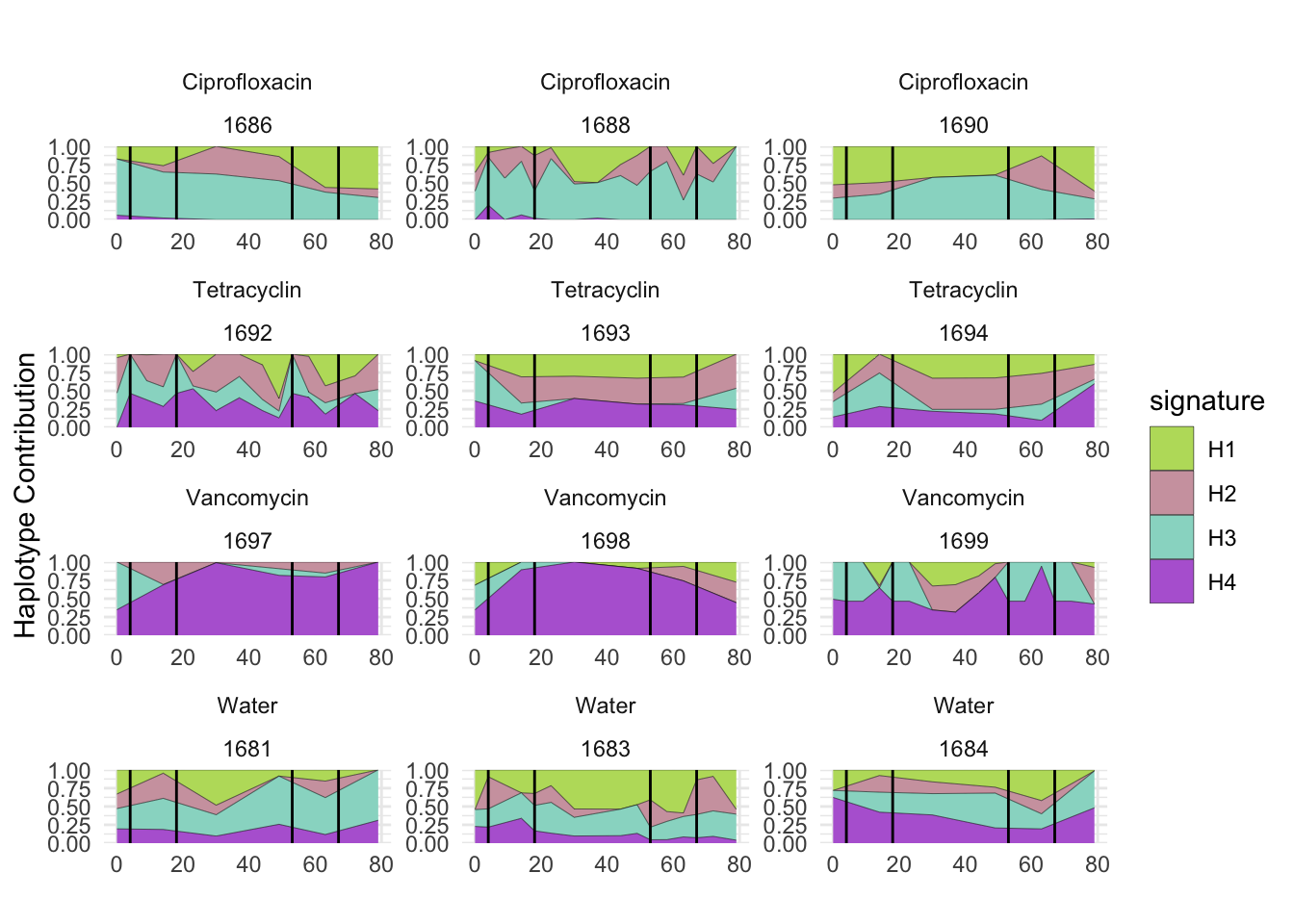
plotSamples(decomposed, normalize = T, remove.sample.names = T, title = bug)
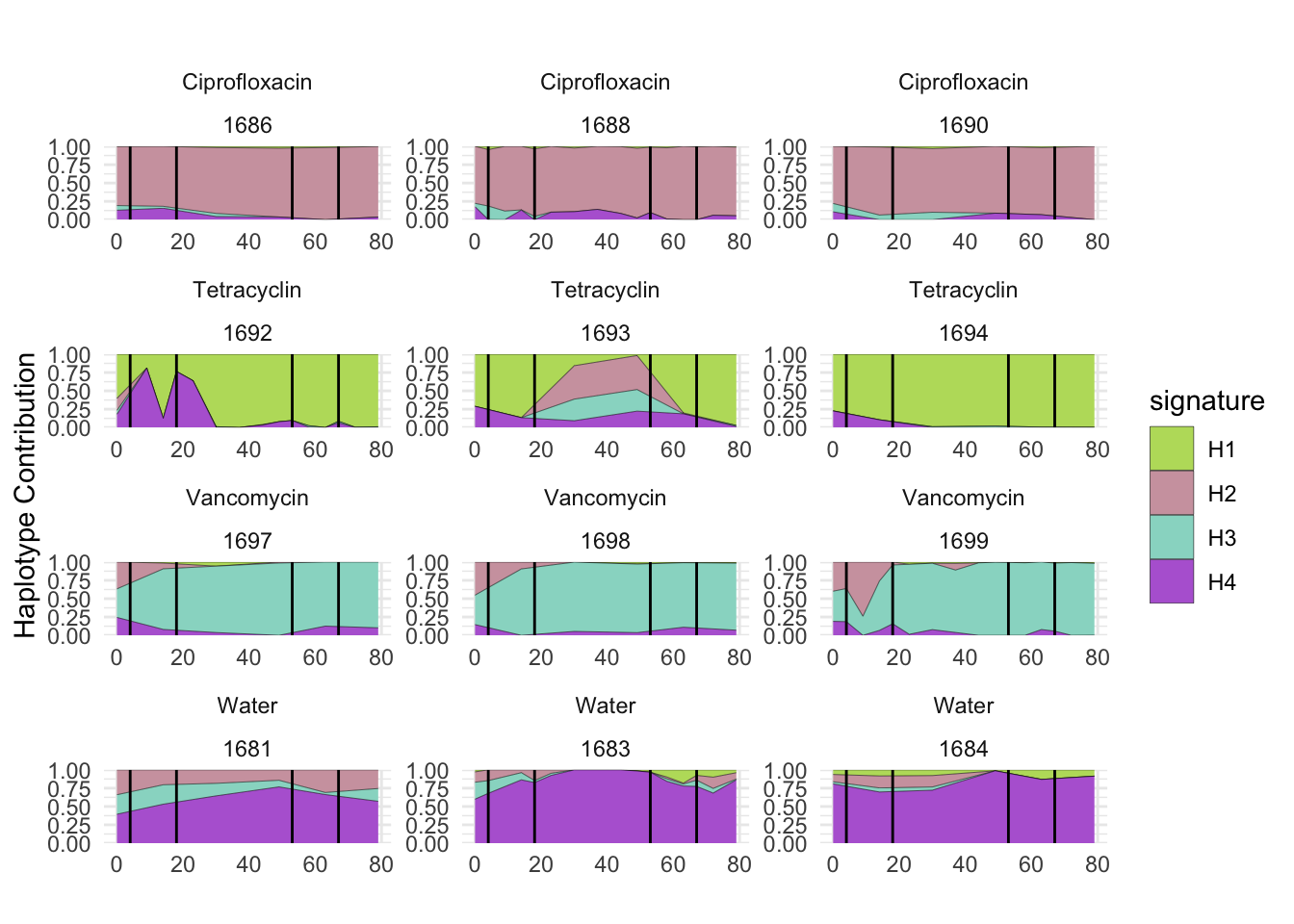
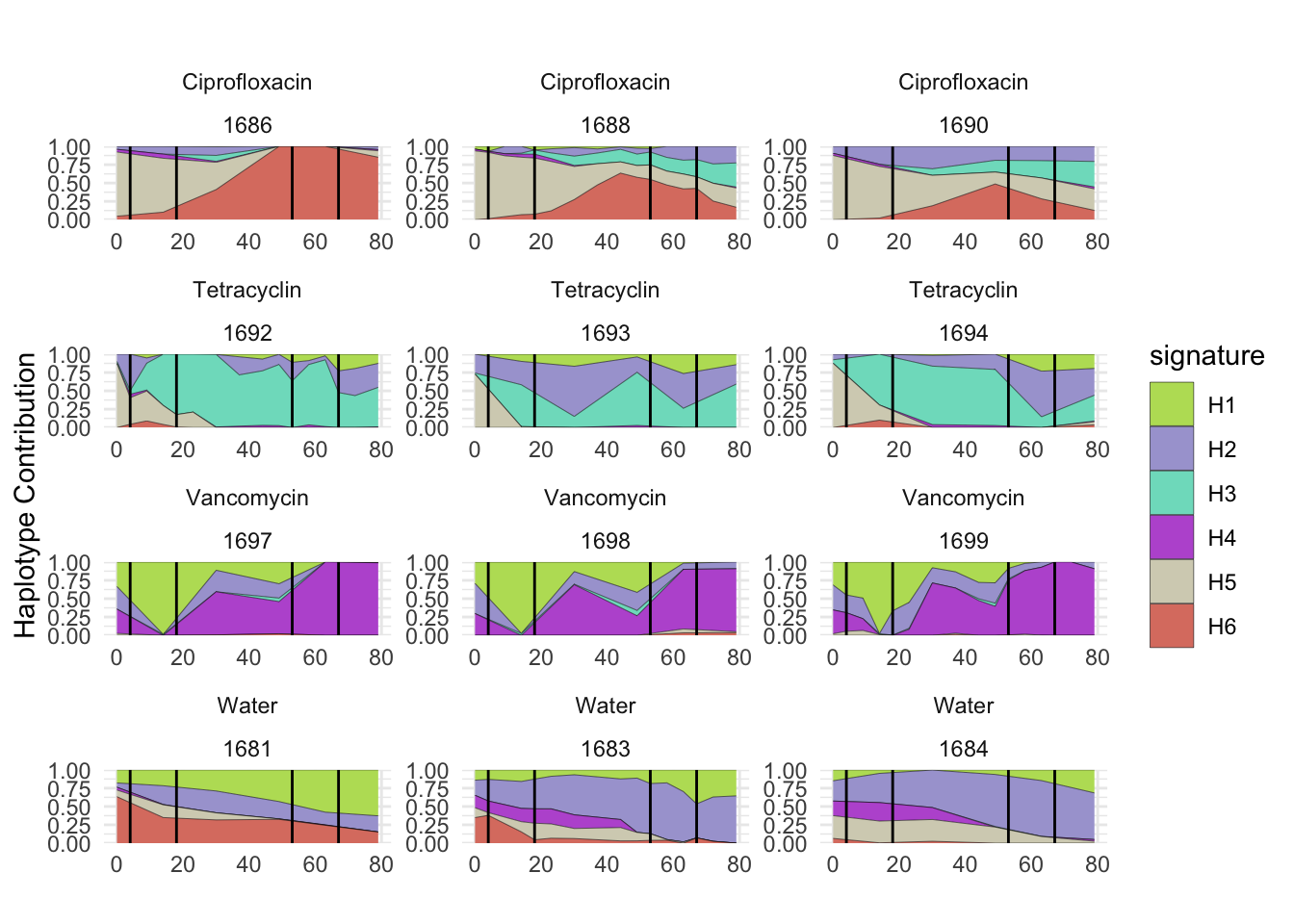
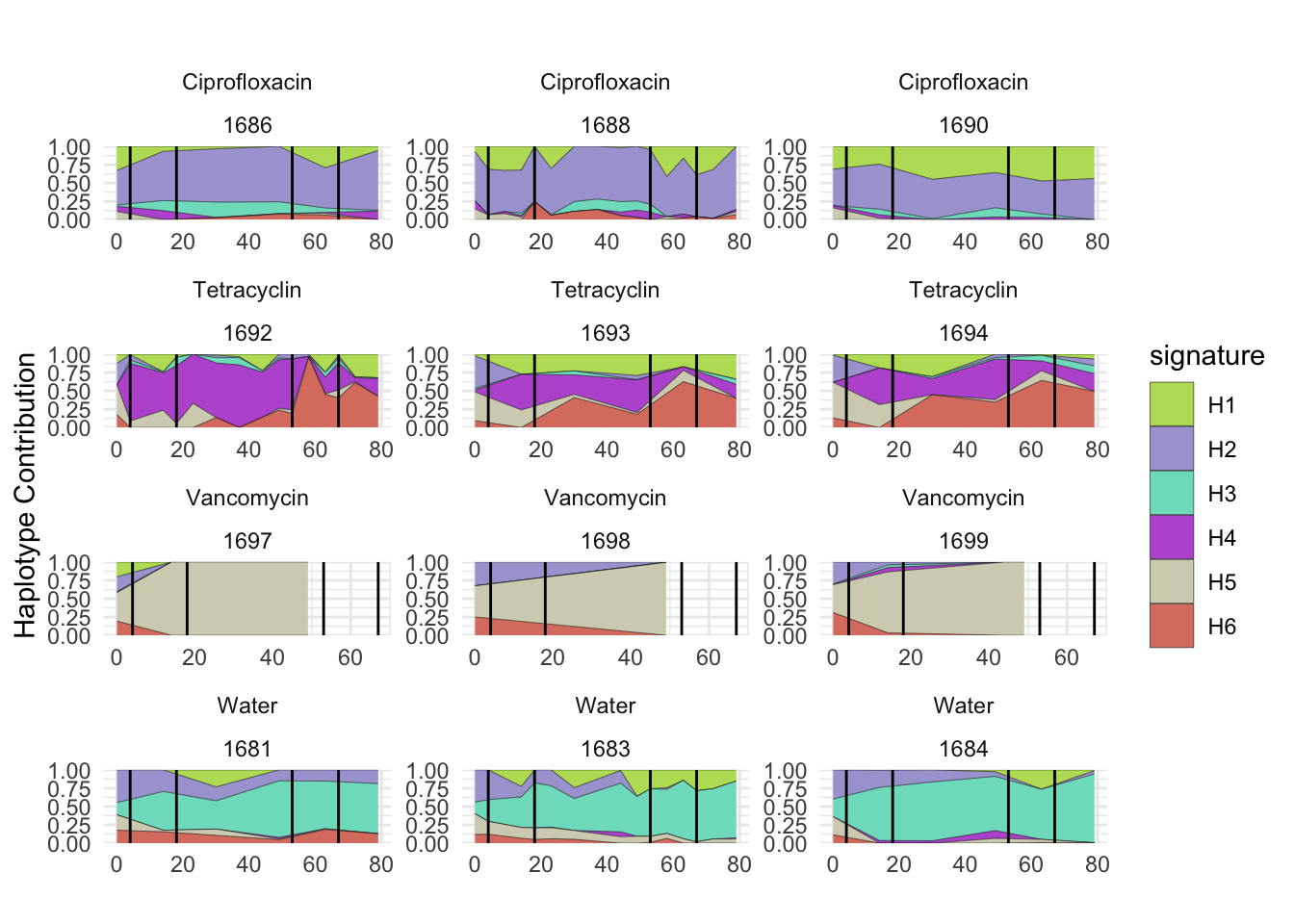
plotSamplesByGroup(decomposed, omm_metadata, normalize = T, title = bug)
plotSamplesByGroup2(decomposed, omm_metadata, normalize = T)
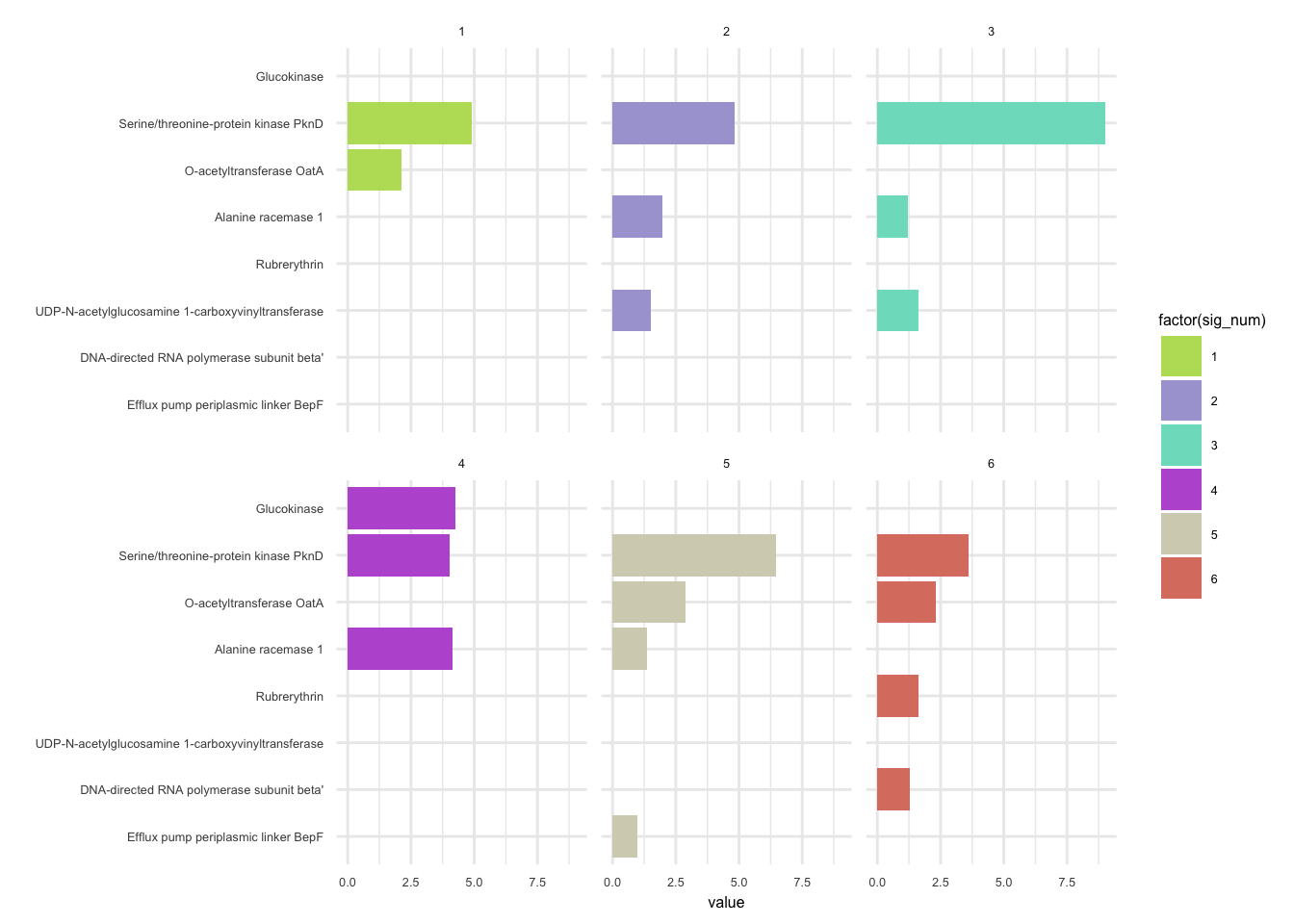
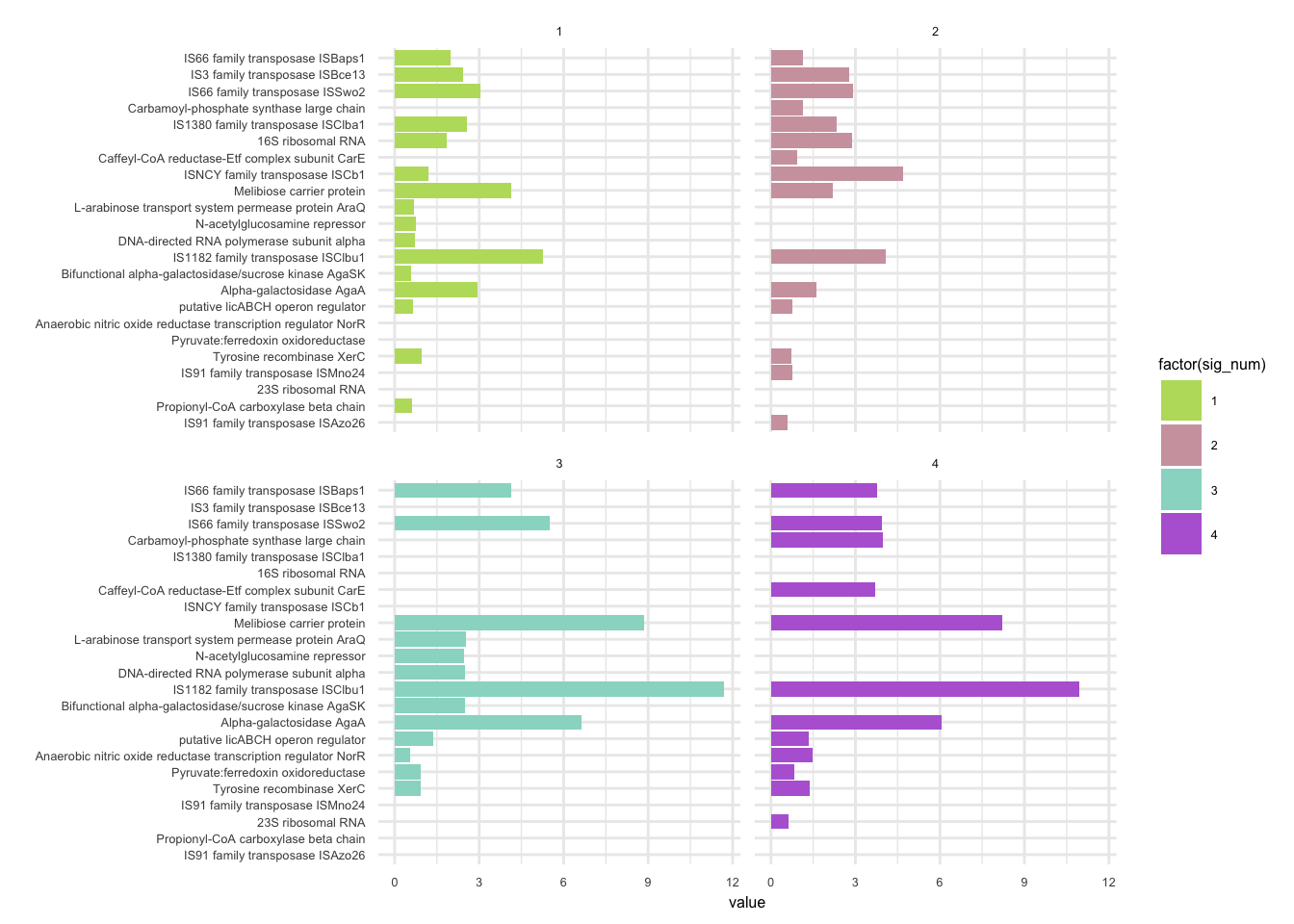
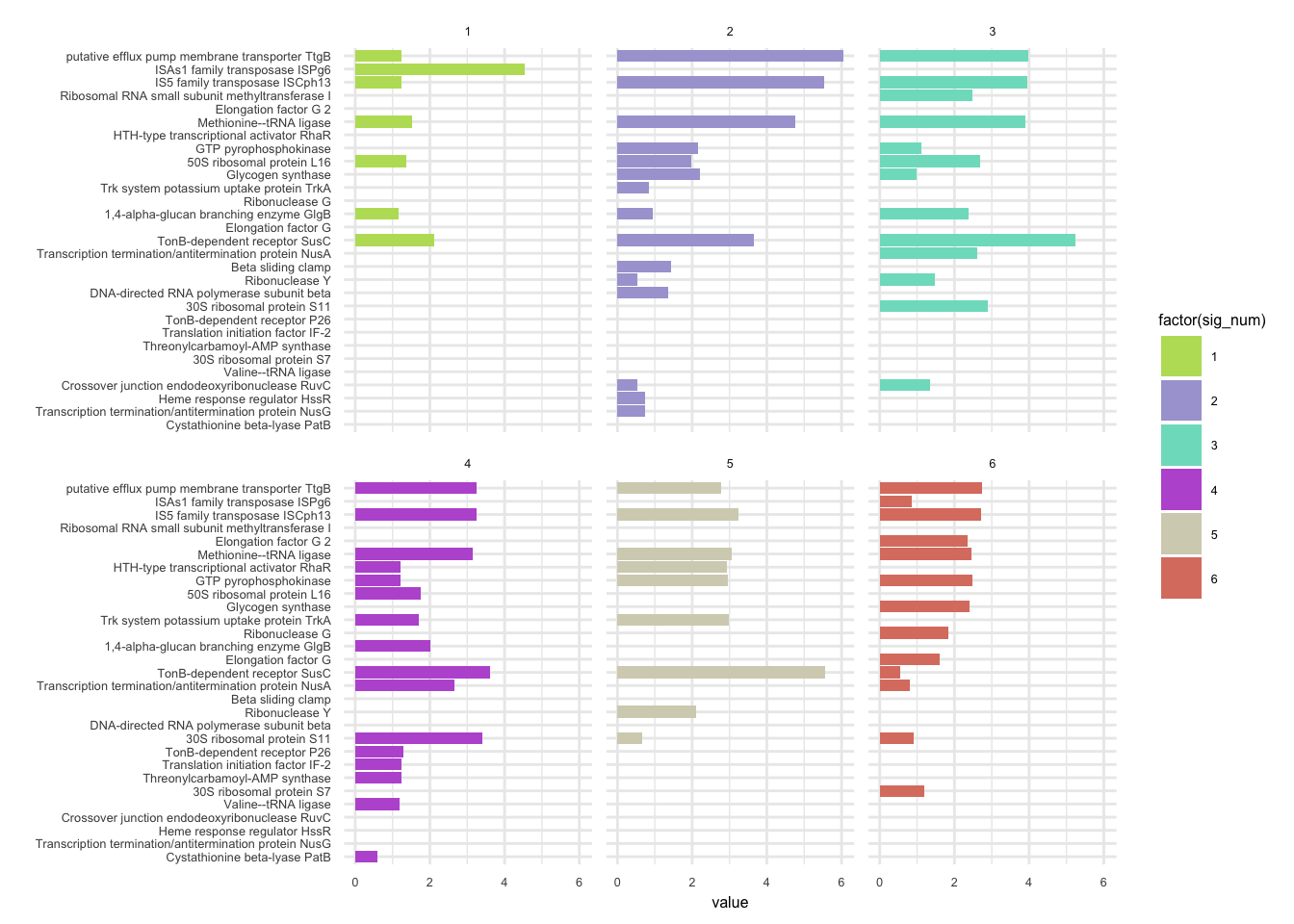
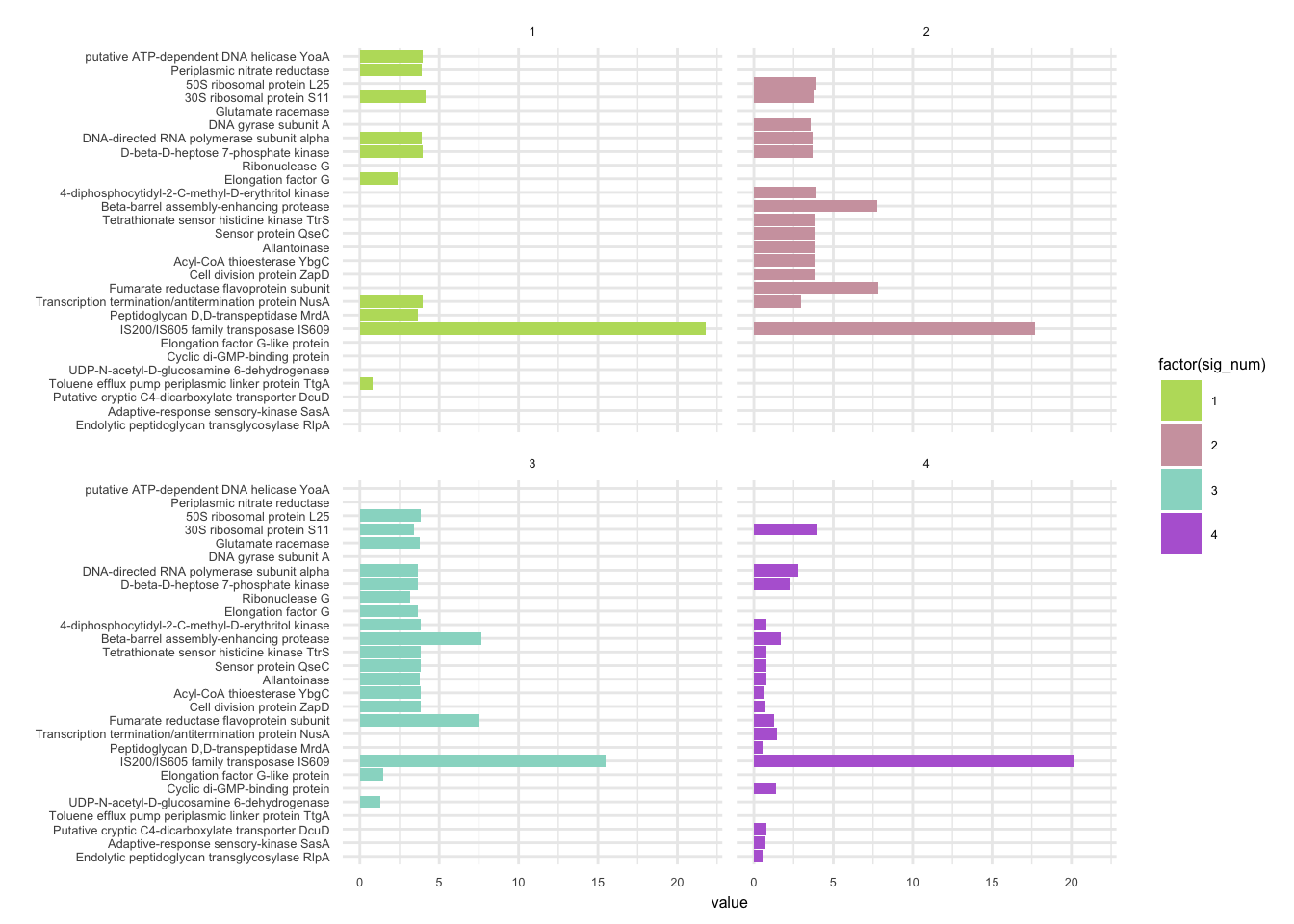
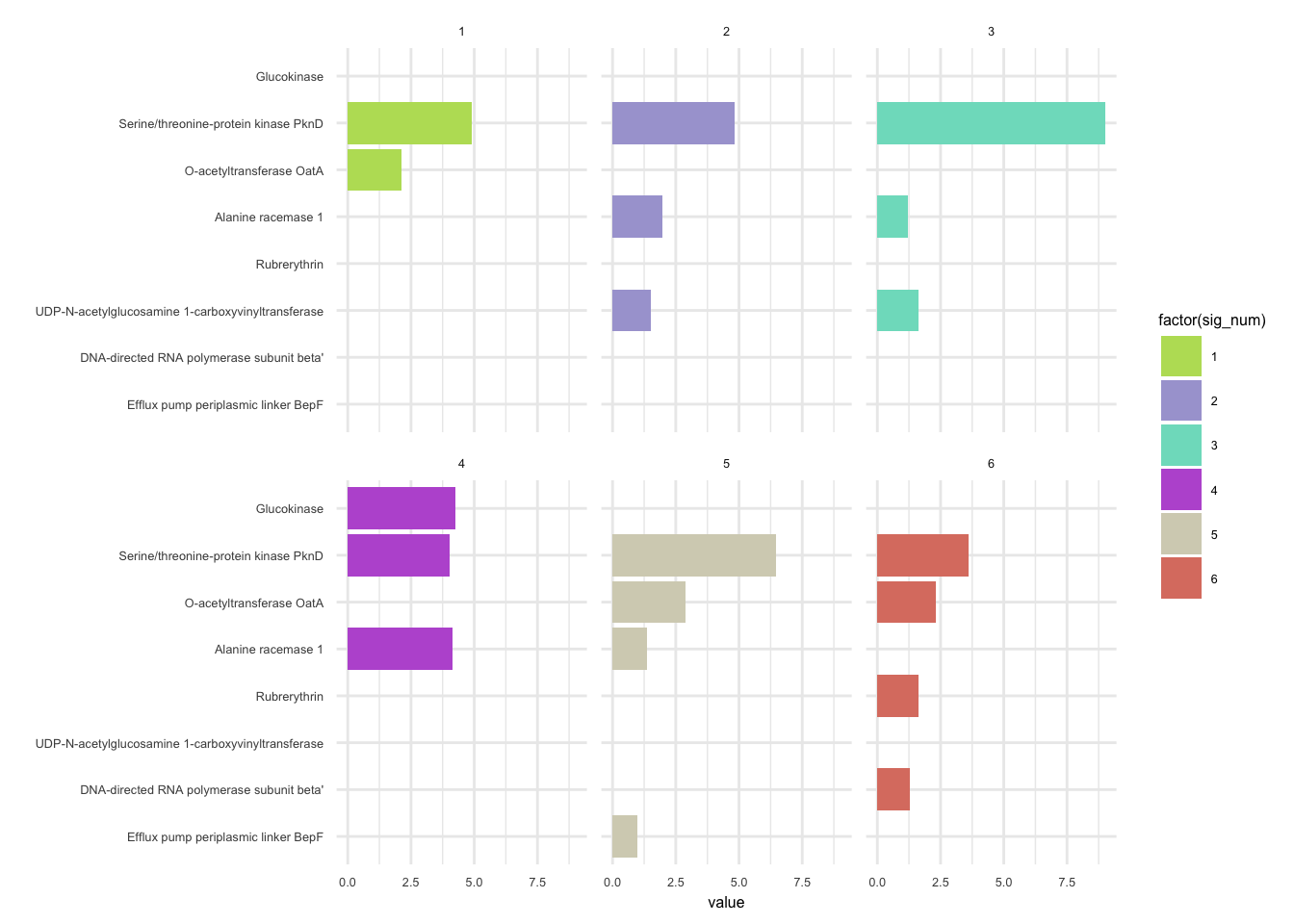
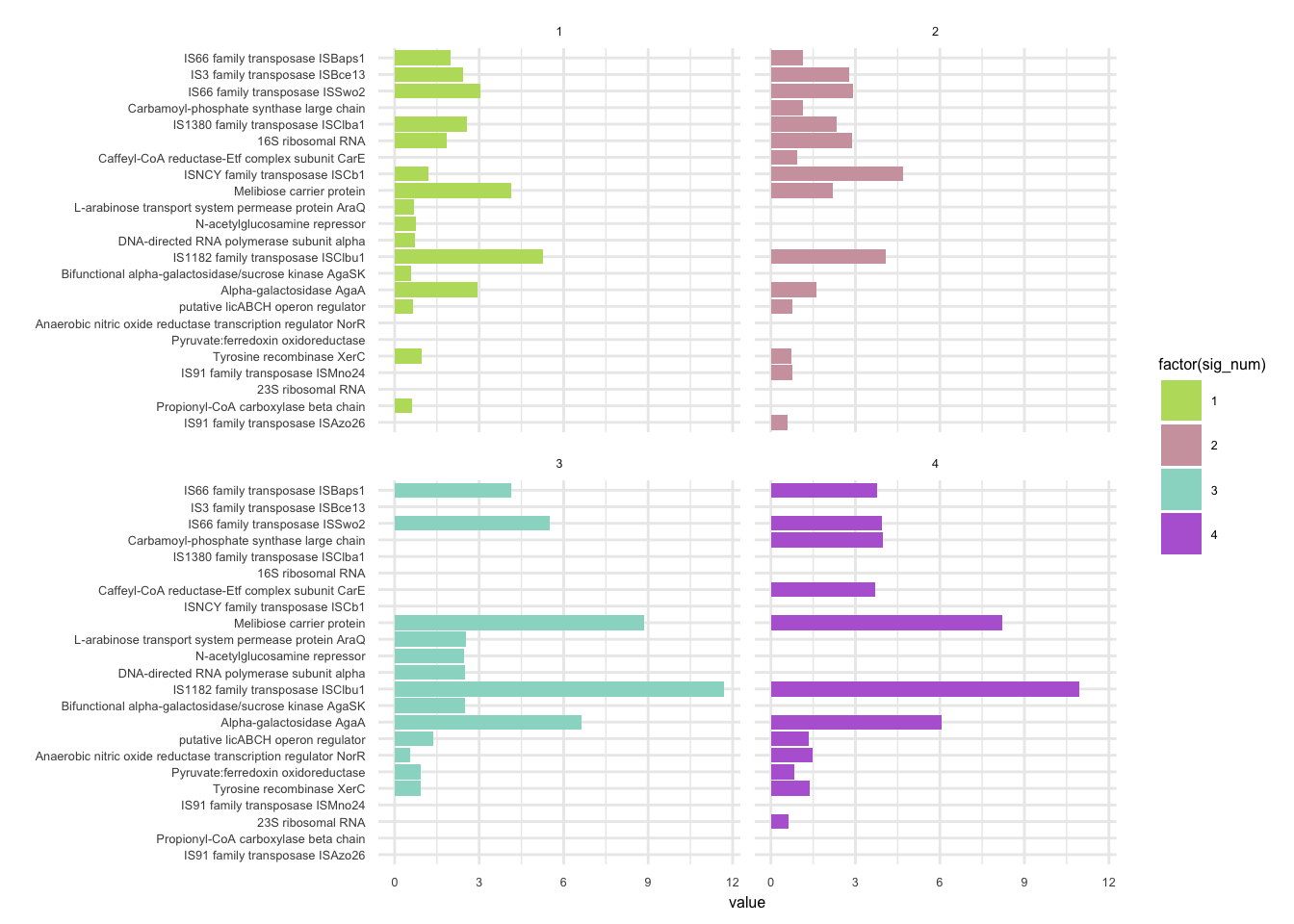
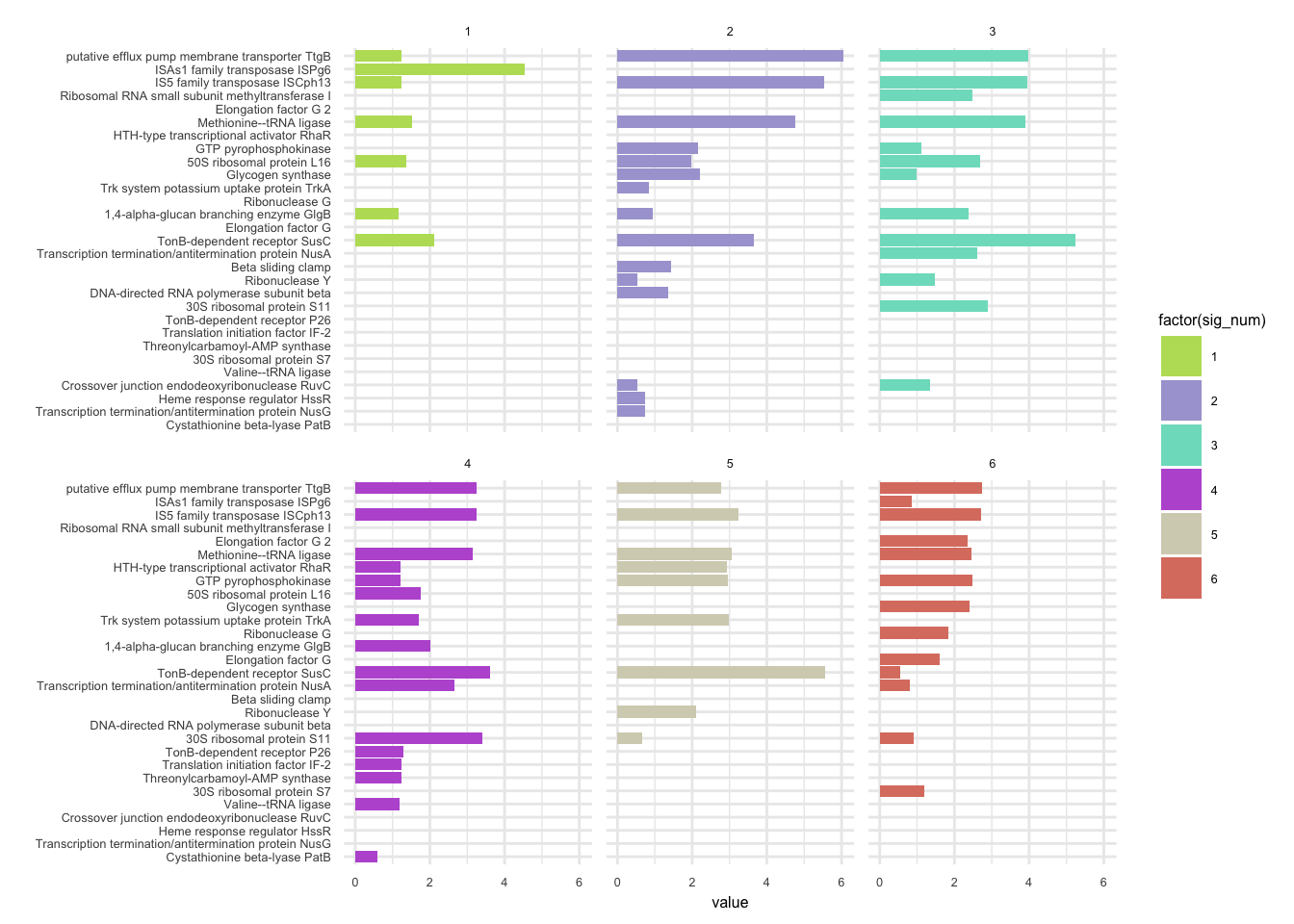
plotHaplotypeAnnotation(decomposed, omm_snp_annotation, hide_hyp = T, sig_threshold = 3)6.10 F_plautii_2
bug <- "F_plautii_2"
omm <- create_df(dat, datre, bug)
# gof <- assessNumberHaplotyes(data.matrix(omm), 2:15, nReplicates = 1)
# plotNumberHaplotyes(gof)
decomposed <- findHaplotypes(data.matrix(omm), 4)
plotHaplotypeMap(decomposed)
plotSamples(decomposed, normalize = F, remove.sample.names = T, title = bug)
plotSamples(decomposed, normalize = T, remove.sample.names = T, title = bug)
plotSamplesByGroup(decomposed, omm_metadata, normalize = T, title = bug)
plotSamplesByGroup2(decomposed, omm_metadata, normalize = T)
# plotHaplotypeAnnotation(decomposed, omm_snp_annotation, hide_hyp = T, sig_threshold =
# 0.0001)